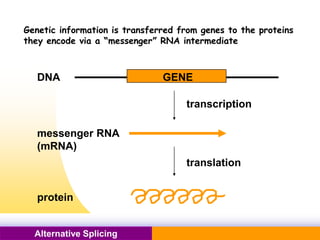
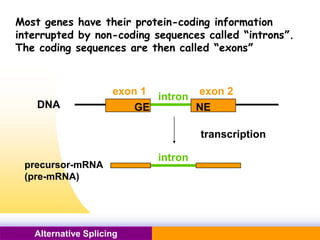
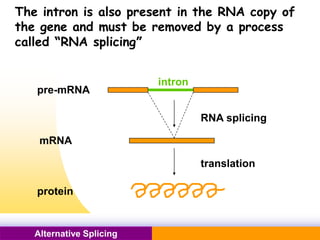
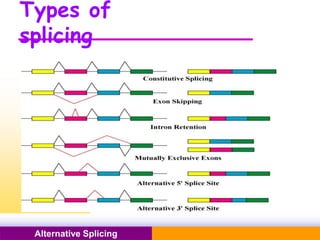
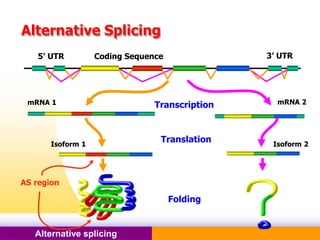
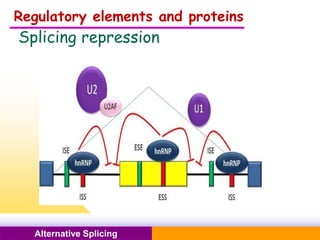
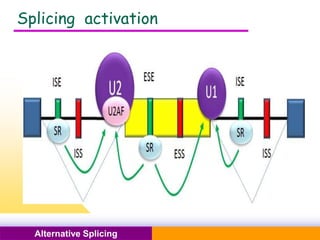
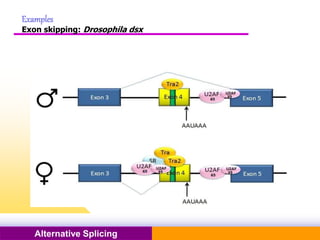
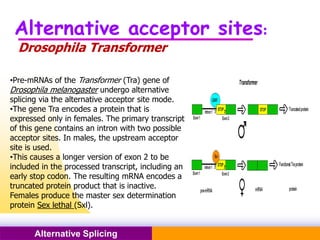
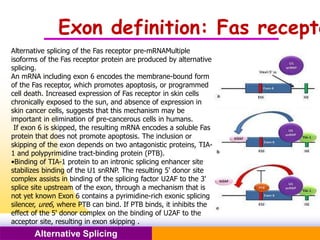
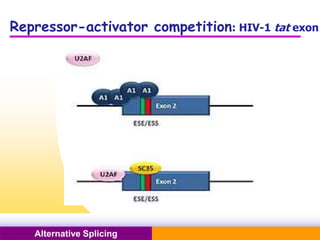
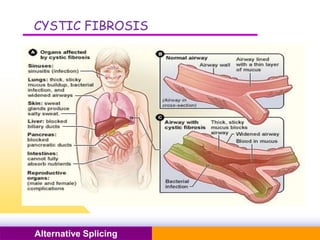
The document discusses alternative splicing, a process by which a single gene can produce multiple protein isoforms through different combinations of exons during RNA splicing. It outlines the mechanisms, regulatory elements, and significance of alternative splicing, highlighting its role in coding variations, evolutionary flexibility, and implications in diseases, particularly cancer. The document also covers the history of its discovery and various examples, including species-specific cases and potential disease linkages.