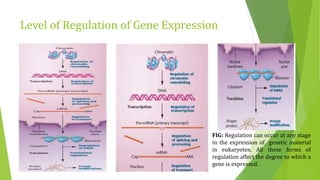
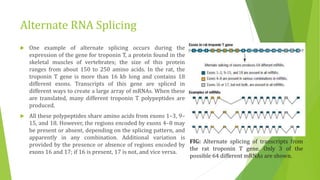
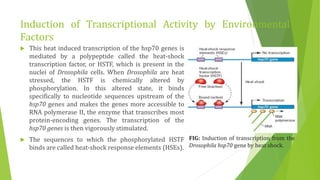
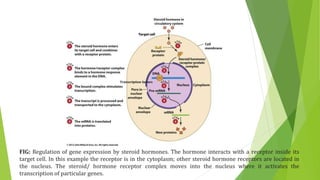
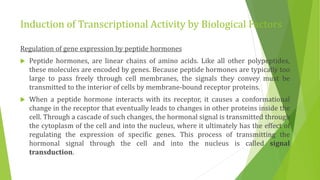
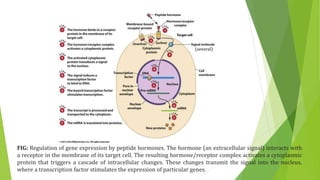
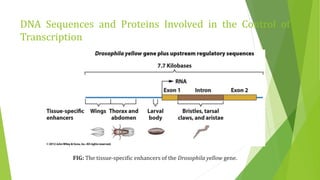
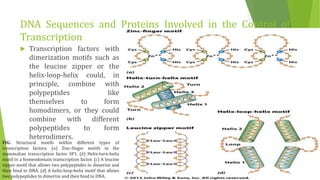
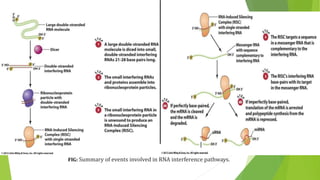
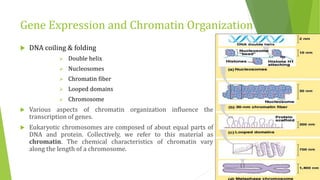
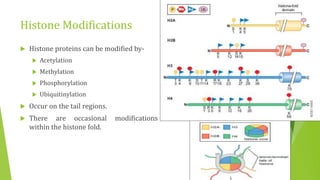
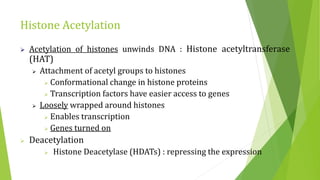
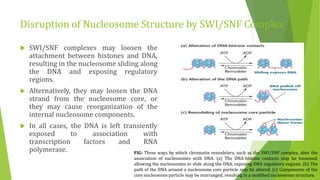
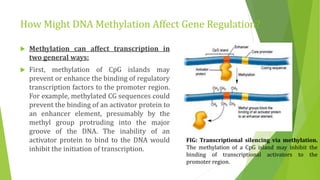
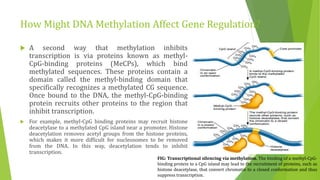
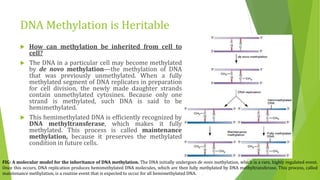
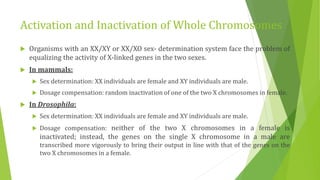
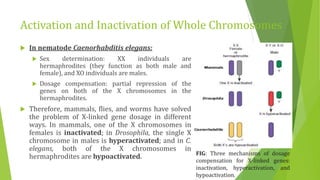
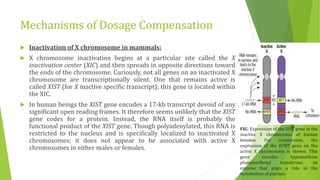
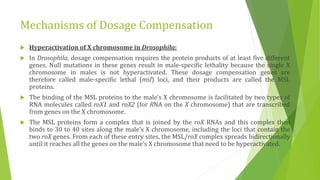
The document discusses various mechanisms of regulating gene expression in eukaryotes. It explains that regulation can occur at multiple levels, including DNA, transcription, mRNA processing, and protein synthesis. Key points include: (1) Regulation allows adaptation and cellular differentiation; (2) In eukaryotes, transcription and translation are separated, allowing more complex regulation; (3) Regulation mechanisms include controlling chromatin structure, transcription initiation, mRNA splicing/stability, and protein modifications. Environmental factors like heat and hormones can also induce gene expression changes through transcription factors.