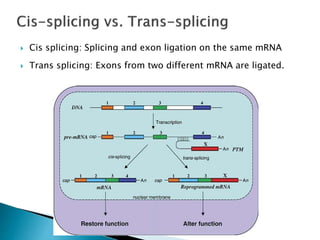
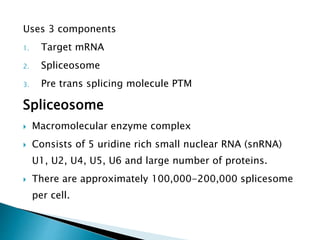
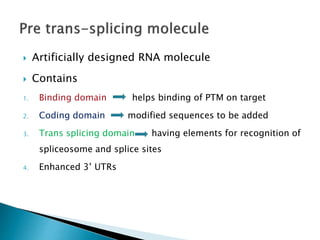
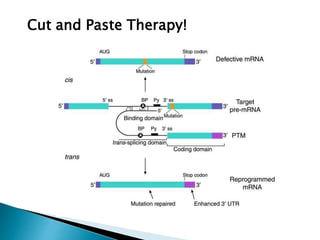
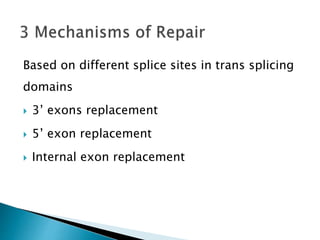
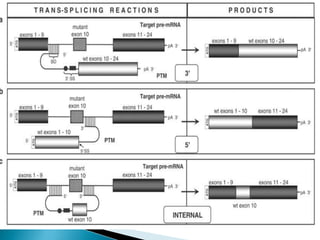
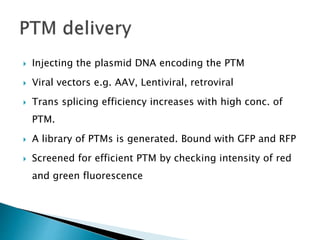
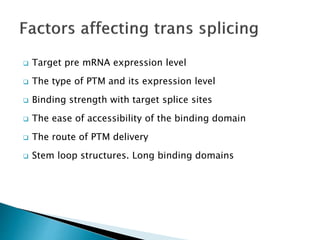
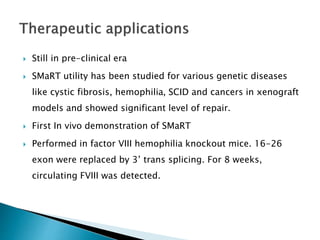
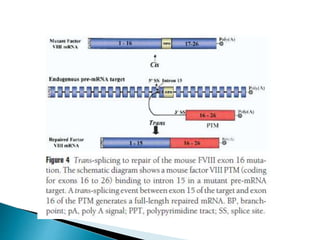
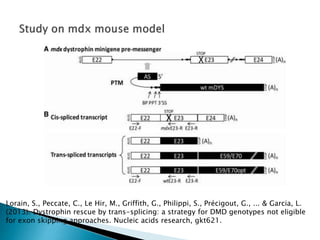
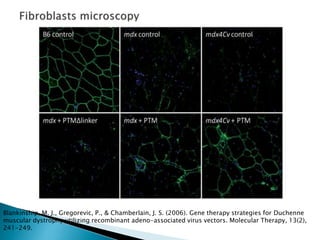
This document summarizes pre-RNA trans-splicing as a gene therapy technique. It involves joining exons from two different primary RNA transcripts end to end through the spliceosome. This emerging technique can be used to repair mutated mRNA by providing an externally designed pre-trans-splicing molecule containing the corrected coding sequence. The technique has been studied for diseases like cystic fibrosis and hemophilia in animal models, showing significant repair. One application is for Duchenne muscular dystrophy, where trans-splicing could replace the mutated dystrophin exon. While still in pre-clinical research, pre-RNA trans-splicing offers targeted repair of mutated genes with potential advantages over other gene therapy approaches.