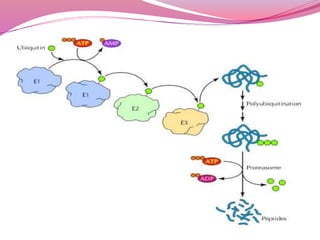
The document discusses protein modifications post-translation, emphasizing their role in protein folding, stability, and localization within cells. Key processes include chaperone-mediated folding, proteolysis, glycosylation, and lipid attachment, all of which are crucial for producing functional proteins from polypeptide chains. Additionally, it highlights the importance of degradation pathways in regulating protein levels and the impact of modifications on protein activity and interactions.

![1-Introduction
2-Protein modifications
[A] Folding
1-Chaperon mediated
2-Enzymatic
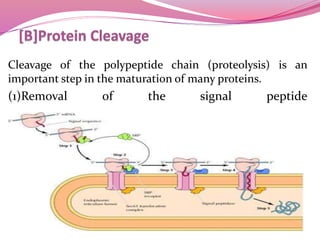
[B] Cleavage
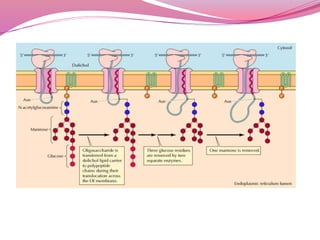
[C] Glycosylation
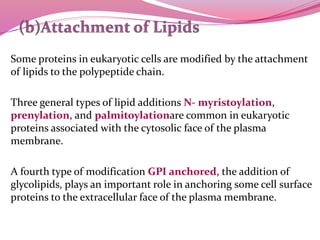
[D] Attachment of Lipids
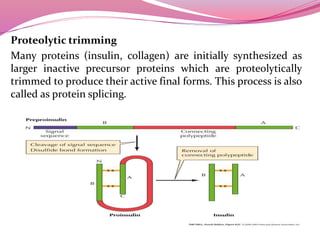
[E] Proteolysis
3-Conclusion
4-Reference](https://image.slidesharecdn.com/2-200527163201/85/co-and-post-translation-modification-by-2-320.jpg)




![Cells contain at least two types of enzymes that act as chaperon.
1 Peptidyl prolyl isomerase [PPI]
2 Protein Disulphide Isomerases (PDI) -
Protein disulfide isomerase or PDI is an enzyme in the ER that
catalyzes the formation and breakage of disulfide
bonds between cysteine residues within proteins as they fold.](https://image.slidesharecdn.com/2-200527163201/85/co-and-post-translation-modification-by-9-320.jpg)