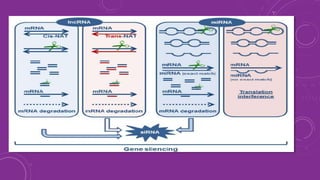
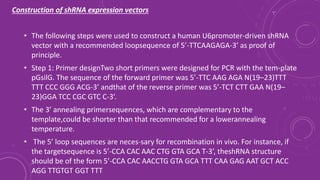
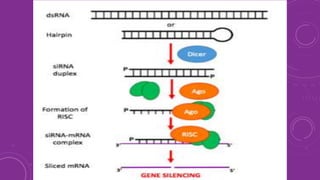
Gene silencing is a process that down regulates gene expression without altering the DNA sequence. It can occur at the transcriptional or post-transcriptional level. Common methods of gene silencing include RNA interference technologies like siRNA, shRNA, and miRNA, which induce degradation of mRNA transcripts. Gene silencing is used for both basic research and applications like crop improvement, as it allows precise down regulation of gene expression.