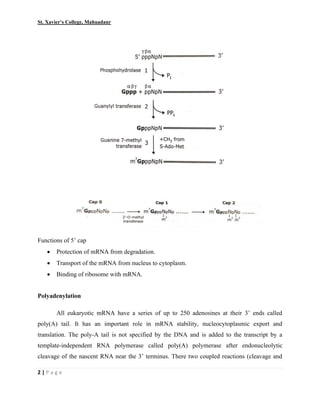
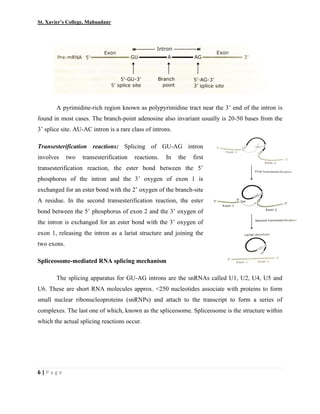
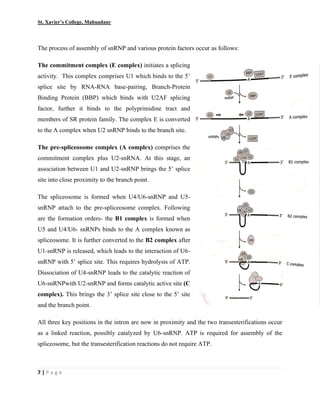
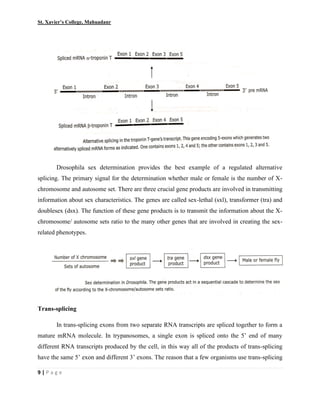
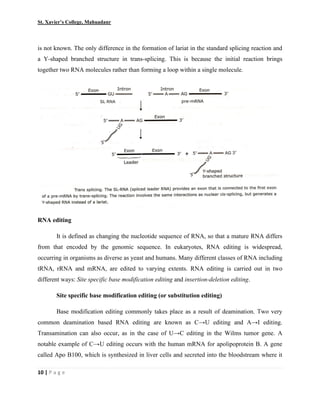
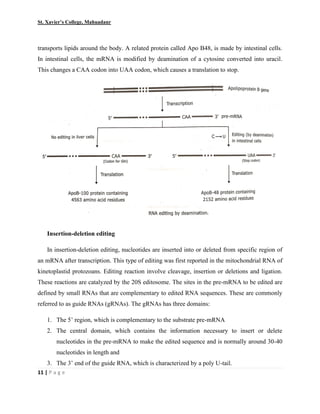
This document summarizes various processes involved in RNA processing in eukaryotes. It discusses 5' capping and polyadenylation of pre-mRNA, as well as splicing of introns and exons. Splicing involves two transesterification reactions catalyzed by a spliceosome complex containing U1, U2, U4, U5 and U6 small nuclear ribonucleoproteins (snRNPs). Alternative splicing and RNA editing are also described, where editing can involve base modification or insertion/deletion of nucleotides guided by small RNAs.