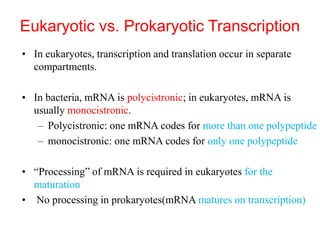
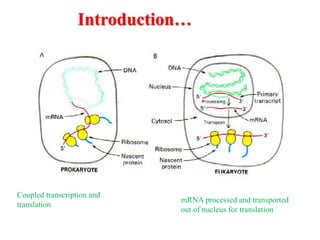
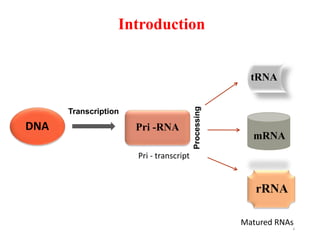
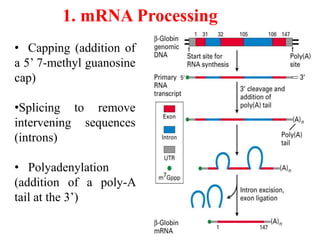
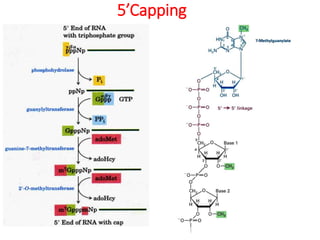
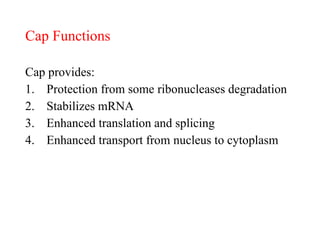
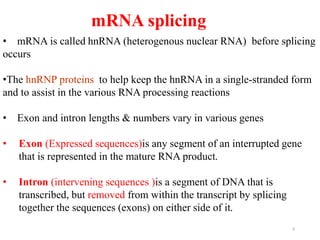
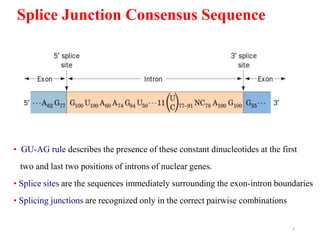
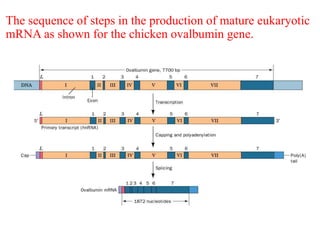
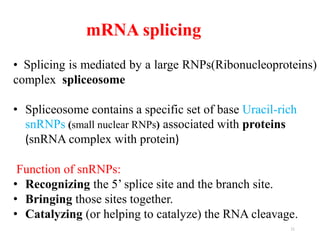
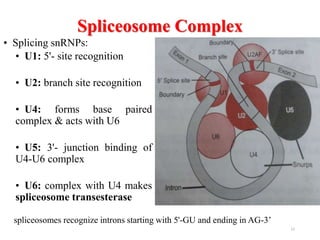
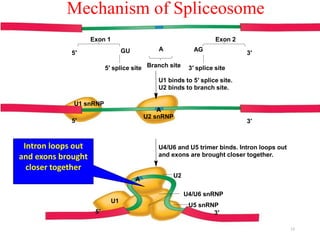
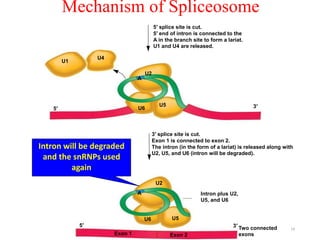
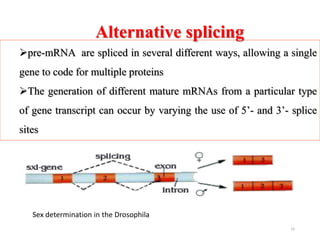
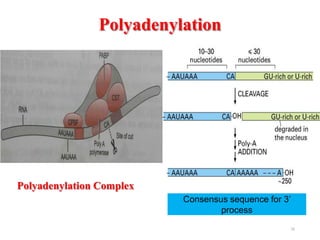
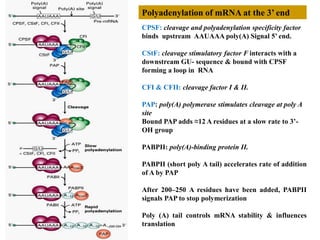
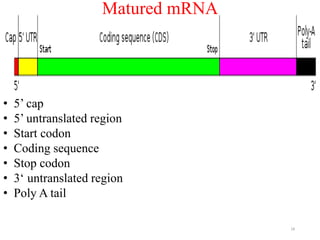
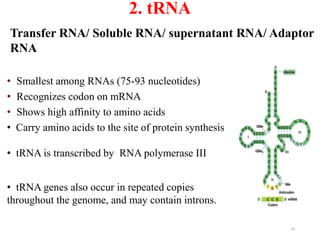
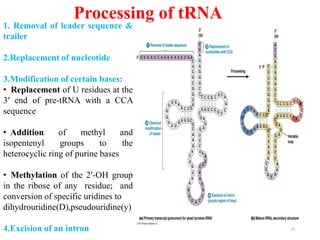
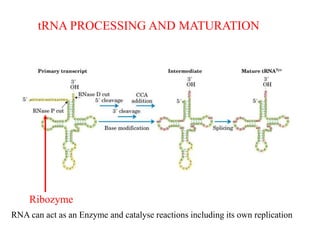
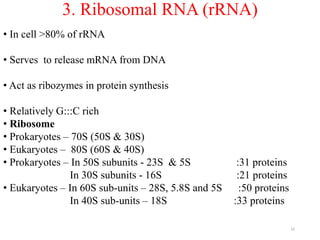
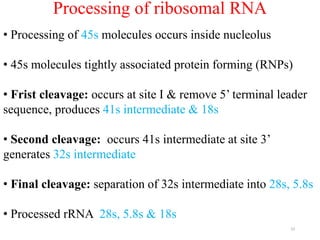
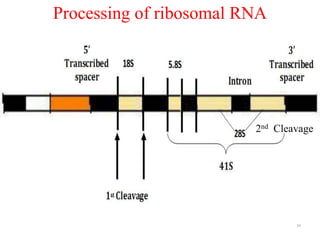
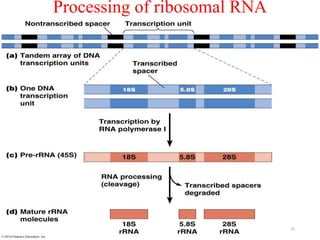
This document summarizes post-transcriptional modifications in eukaryotes. It discusses how eukaryotic mRNA undergoes processing, including capping, splicing to remove introns, and polyadenylation. Splicing requires snRNPs and the spliceosome to recognize splice sites. Alternative splicing allows one gene to code for multiple proteins. tRNA and rRNA also undergo processing as they mature, including modification of bases and removal of sequences. Final mature mRNA, tRNA, and rRNA are then ready for translation.