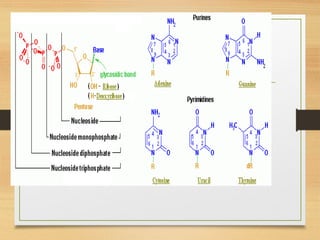
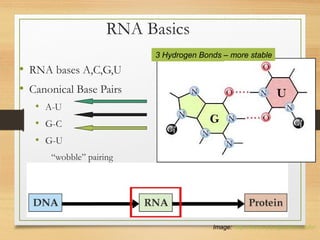
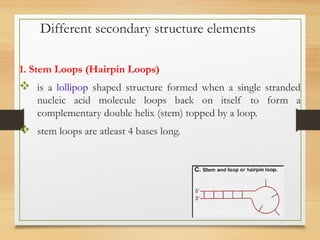
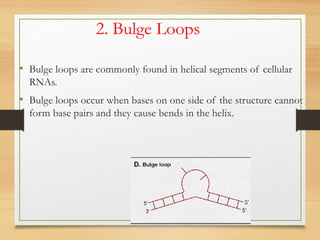
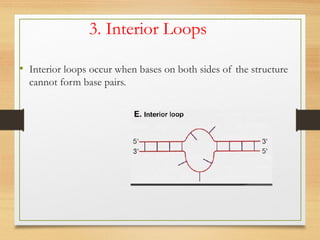
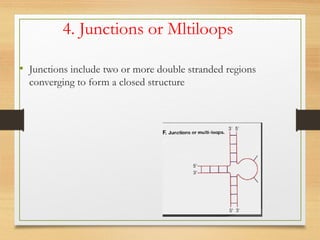
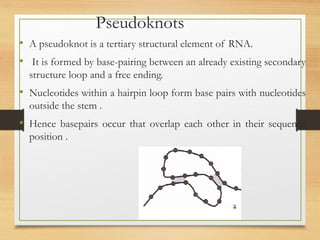
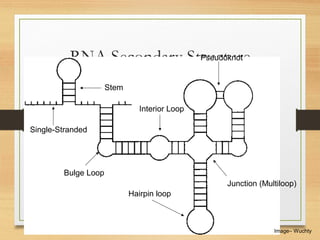
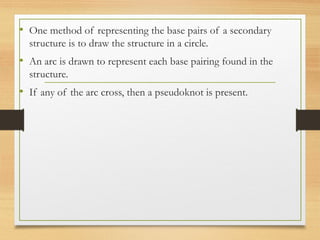
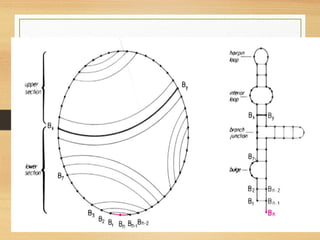
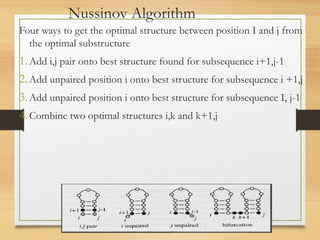
RNA plays an important role in protein synthesis. It has a primary sequence made of A, C, G, and U nucleotides that can fold into a secondary structure through base pairing. The secondary structure is predicted using either maximizing the number of base pairs or minimizing the free energy. Dynamic programming is commonly used to predict the optimal secondary structure. Predicting RNA structure is important for understanding its function and evolution.