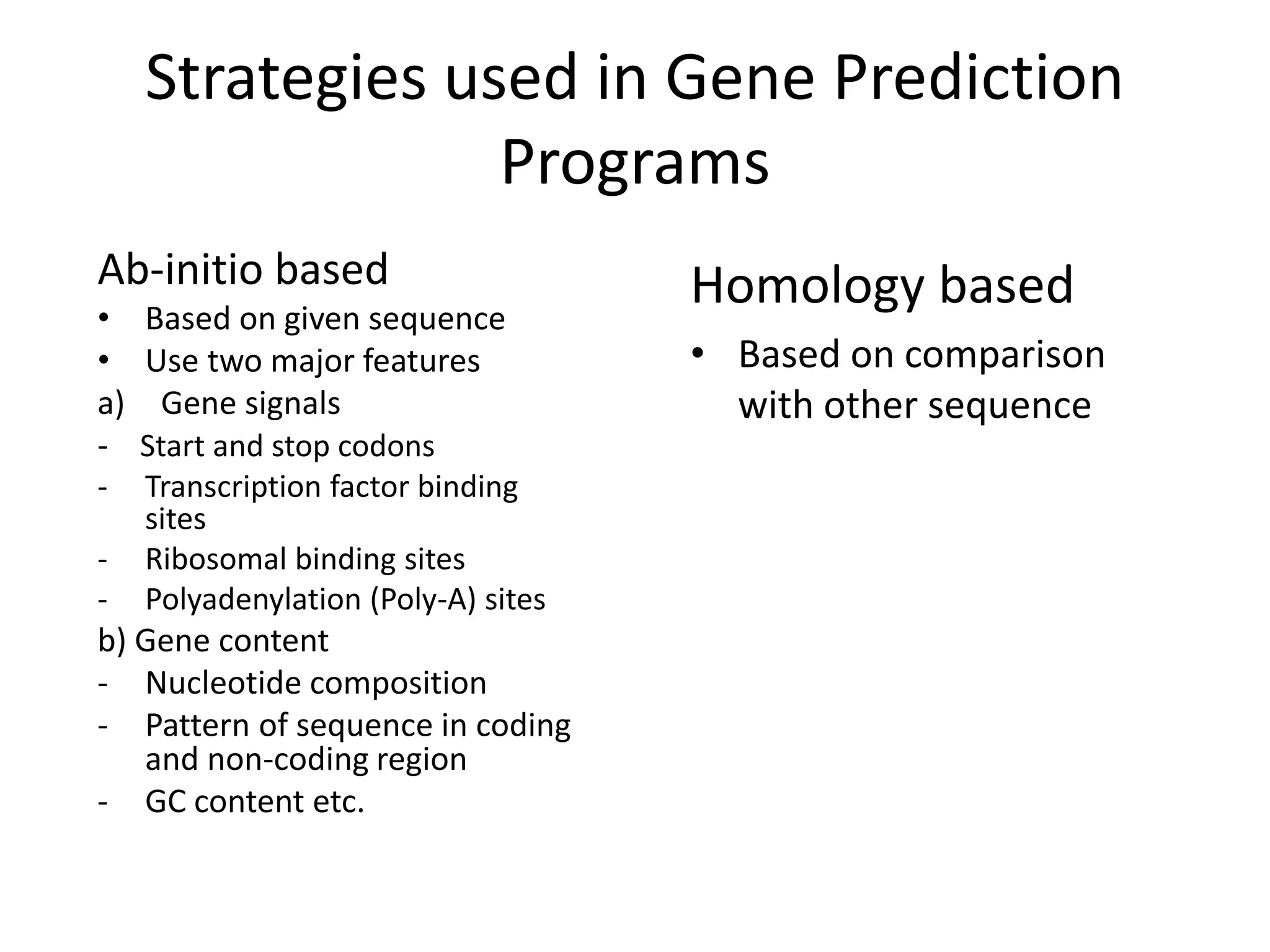
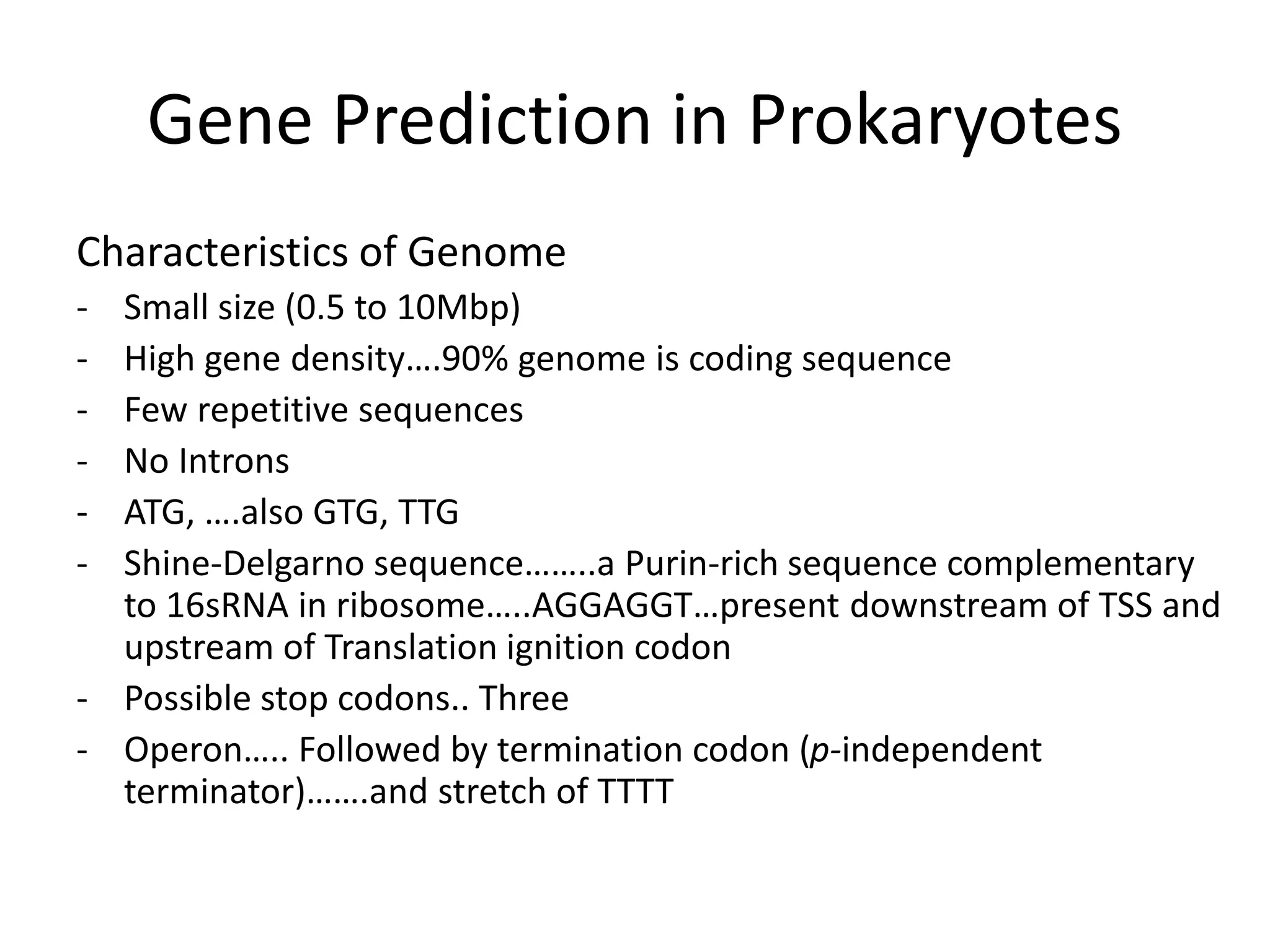
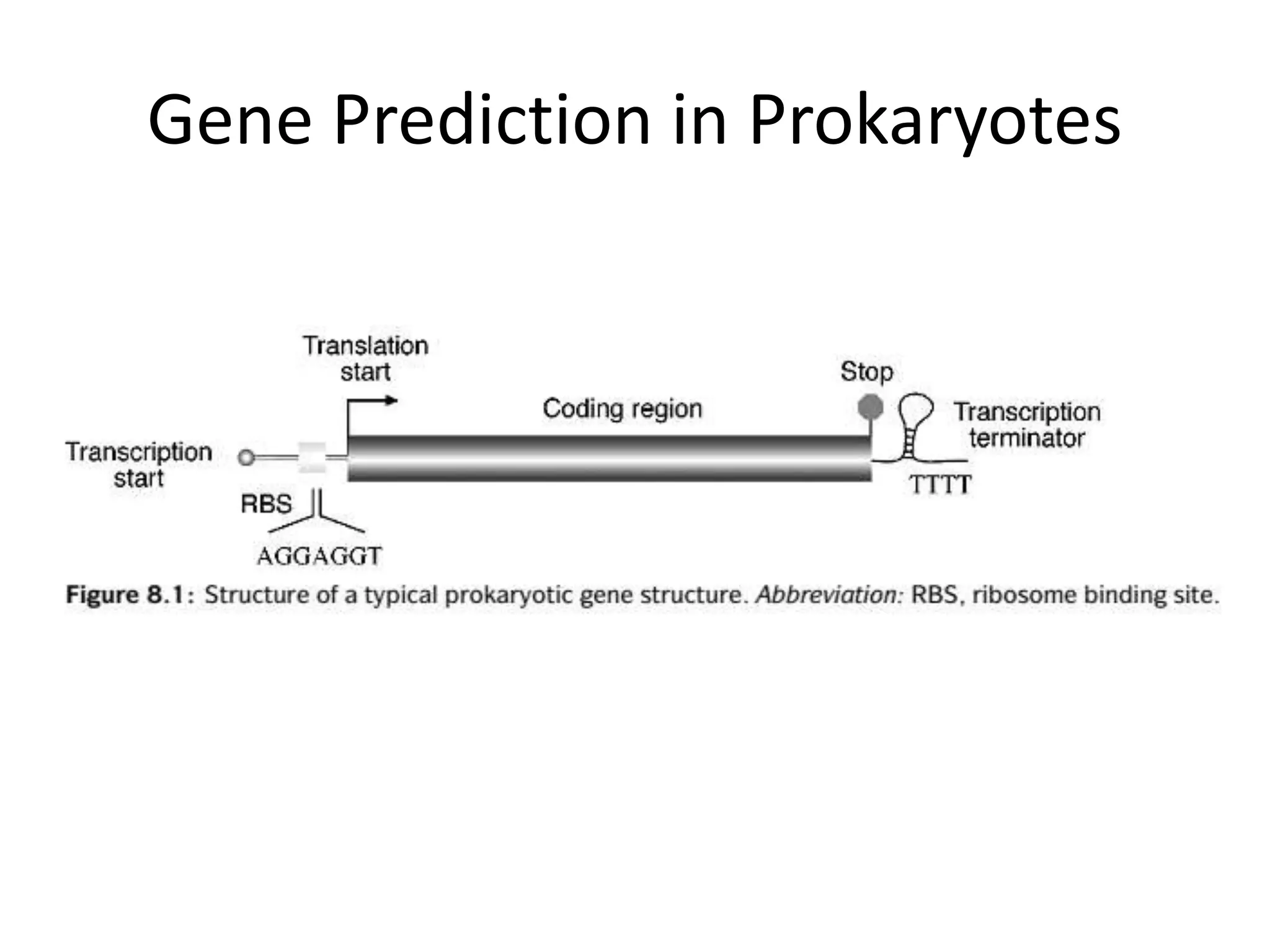
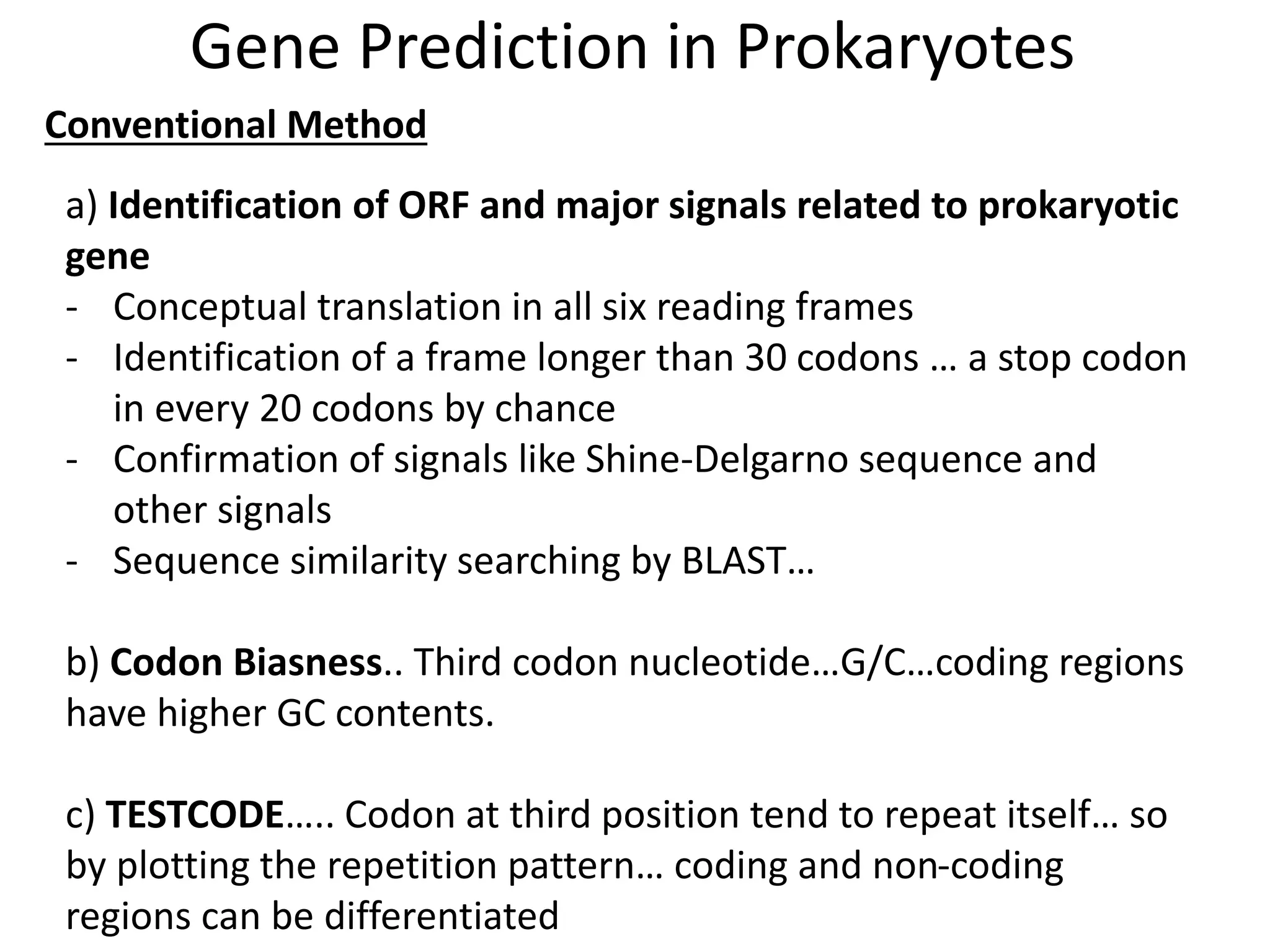
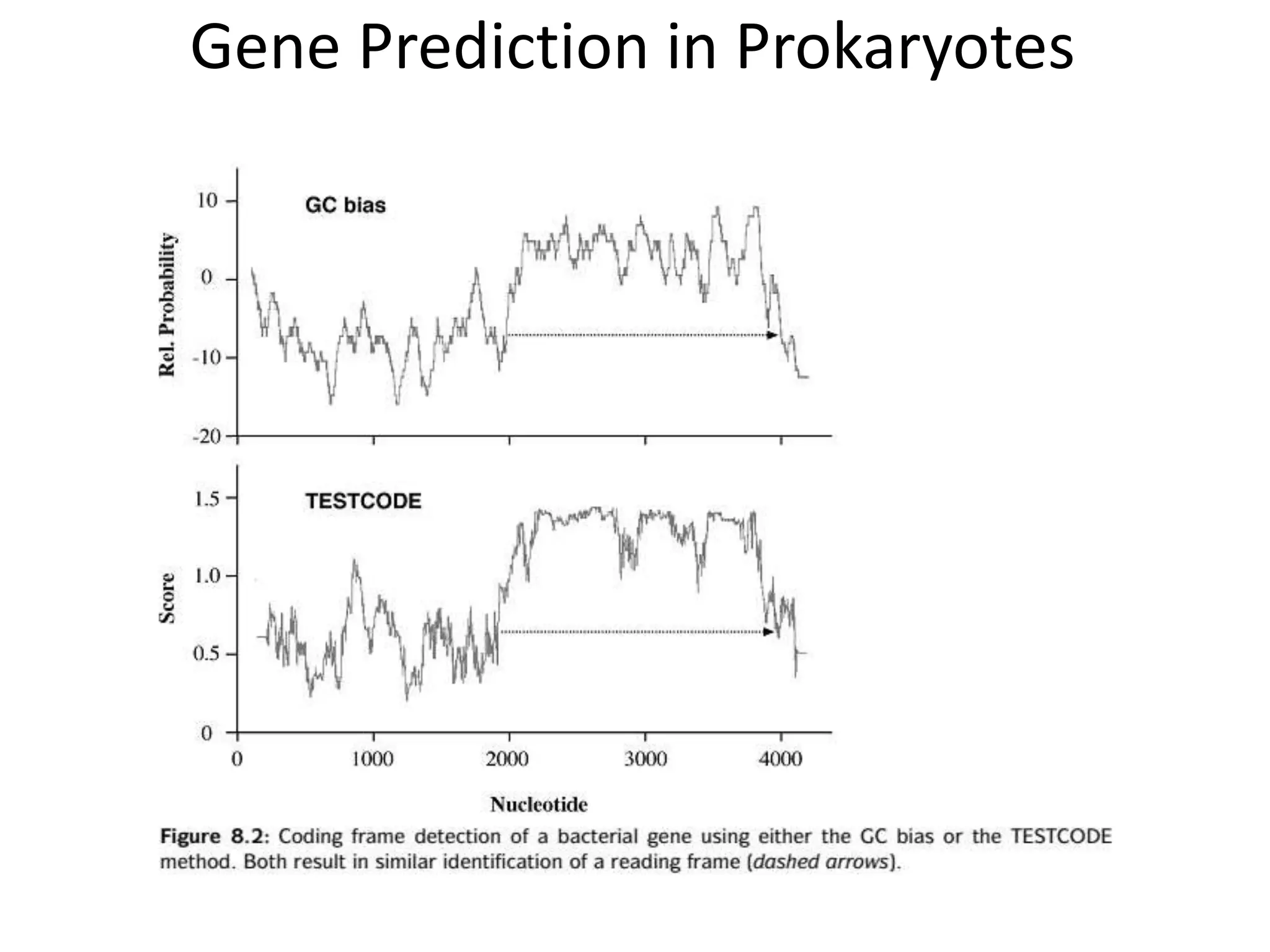
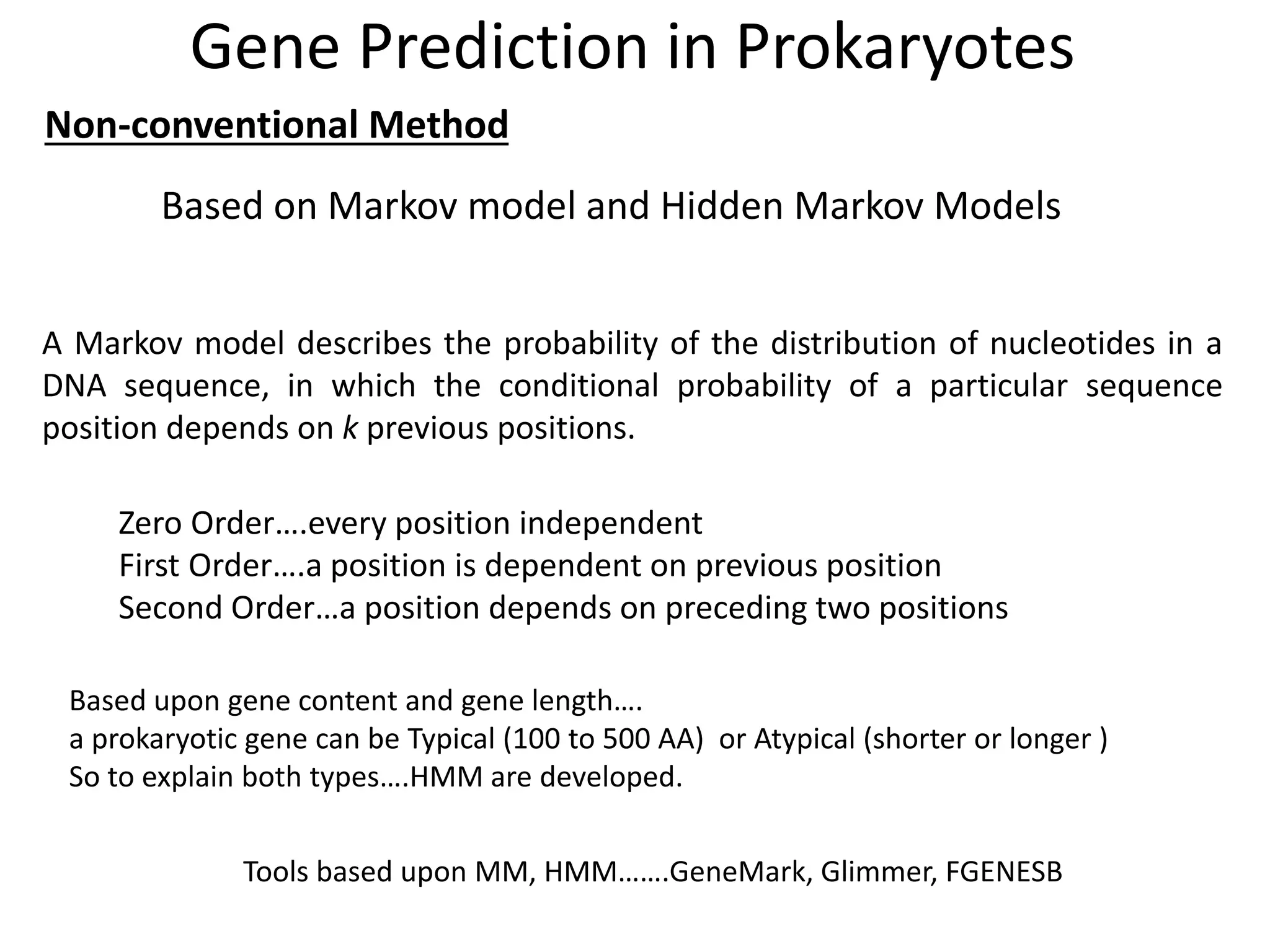
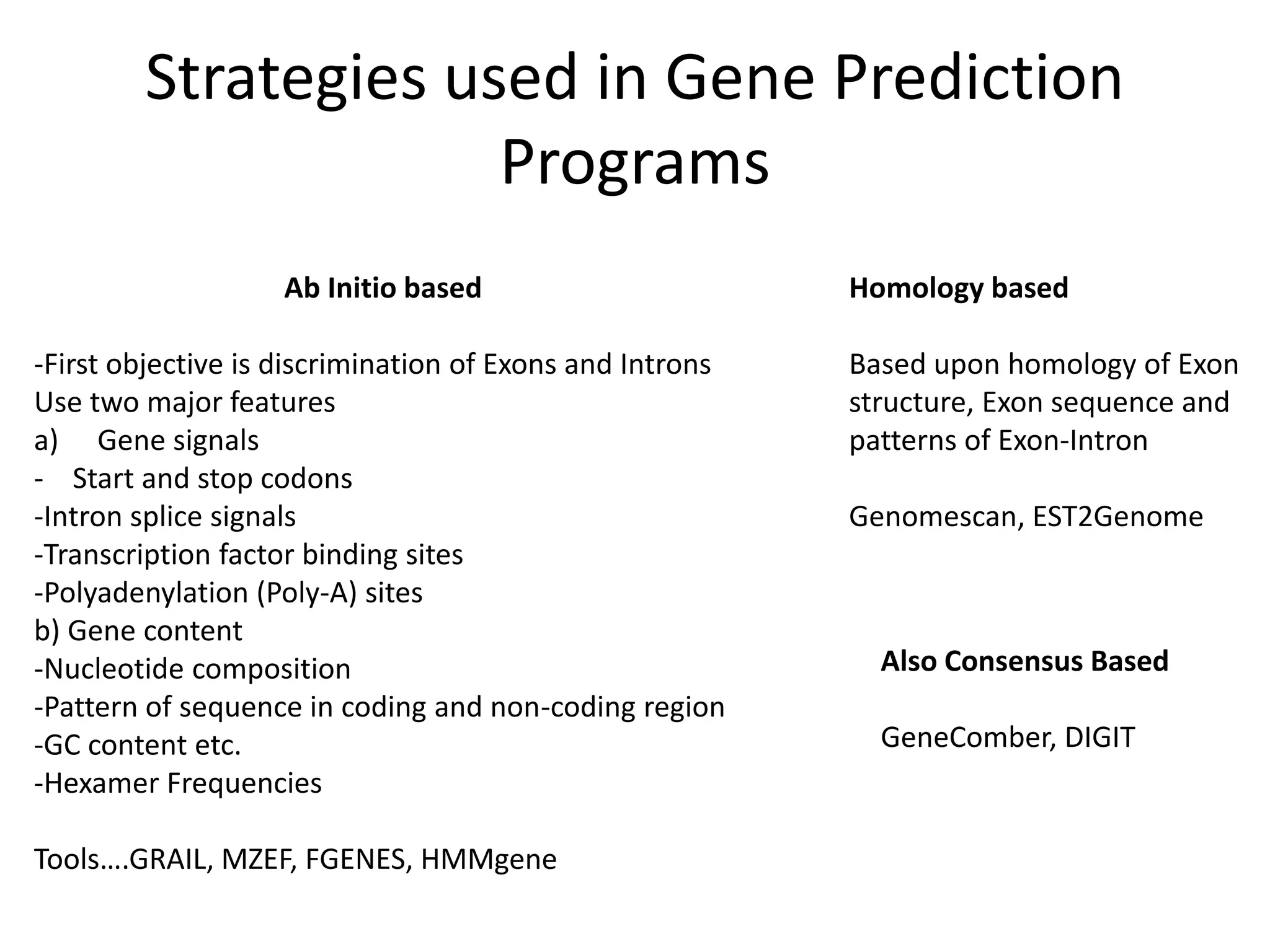
This document discusses strategies for gene prediction in prokaryotes and eukaryotes. It explains that gene prediction programs can be homology-based, using comparisons to other sequences, or ab initio-based, using features like start/stop codons and transcription factor binding sites. For prokaryotes, it outlines characteristics like short genome size and few introns, and conventional methods involve identifying ORFs and signals while newer methods use Markov and hidden Markov models. For eukaryotes, it notes large genome size, many introns and repeats, and the need to distinguish exons from introns using signals and composition patterns. Specific gene prediction tools are also mentioned.