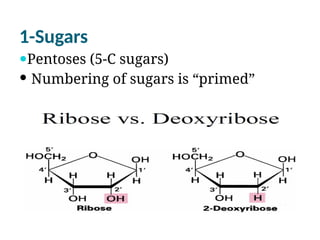
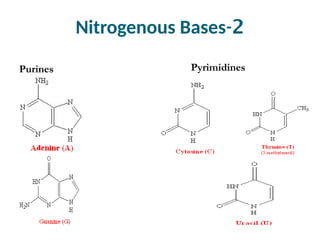
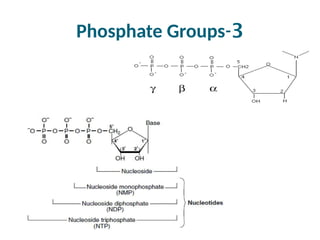
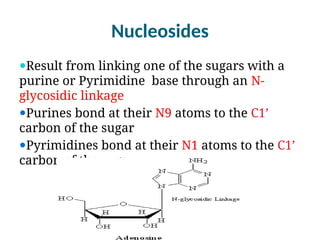
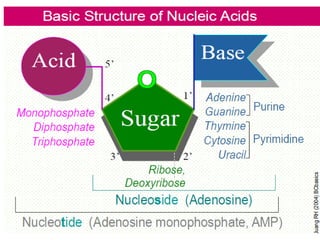
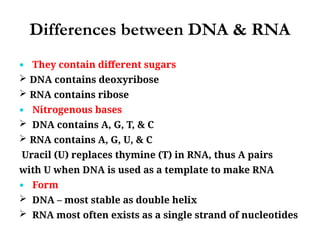
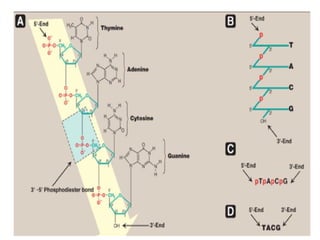
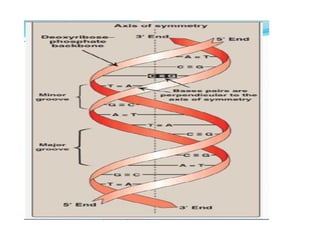
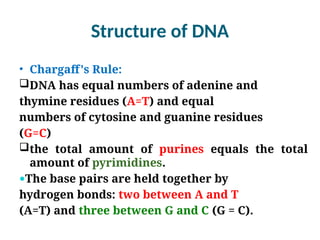
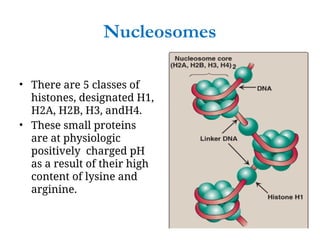
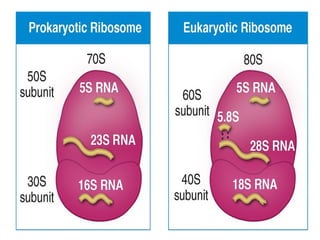
The document discusses nucleic acids, including the structures and differences between DNA and RNA, highlighting their roles in storing and expressing genetic information. It explains nucleotide composition, the double helix structure of DNA, base pairing rules, and the organization of DNA within cells. Additionally, it covers ribosomal RNA and its significance in protein synthesis.