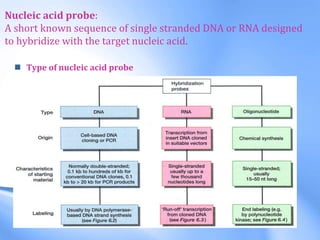
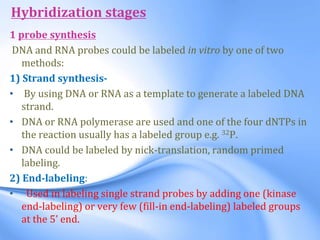
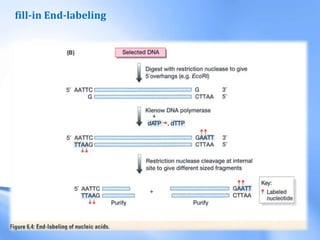
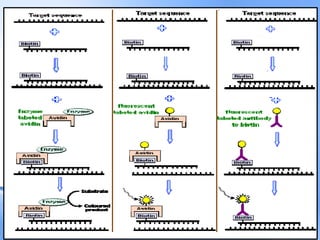
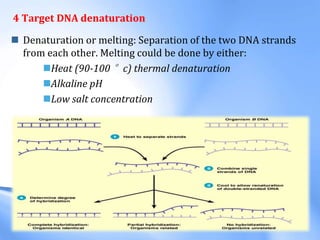
The document discusses nucleic acid hybridization, a key technique in molecular genetics for detecting DNA or RNA sequences through the formation of double-stranded molecules. It outlines the stages of hybridization, including probe synthesis, target DNA processing, denaturation, and methods of transferring target DNA to solid carriers. Various applications of hybridization such as Southern, Northern, and Western blotting are explained, along with the visualization techniques used for identifying hybridized sequences.