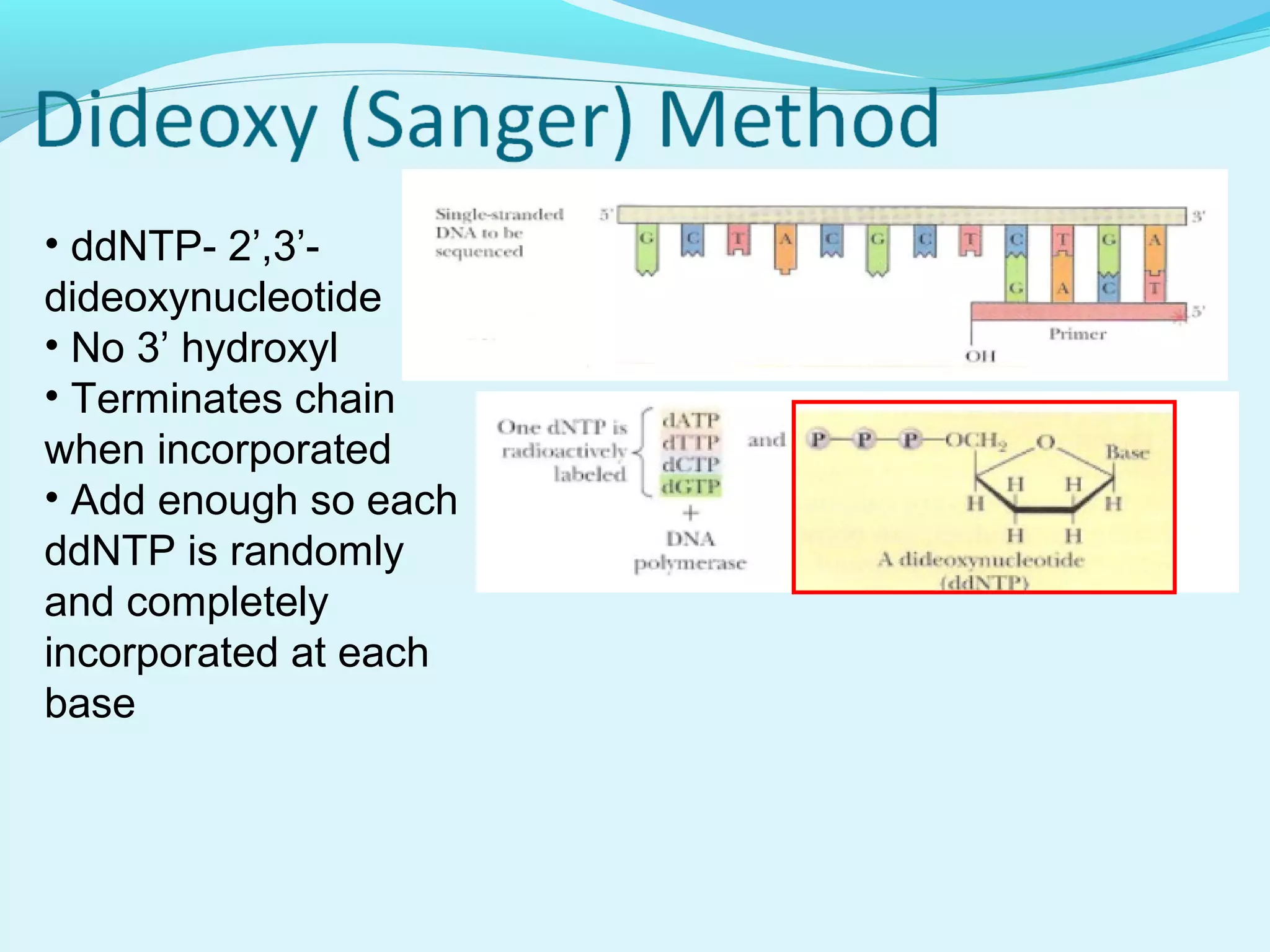
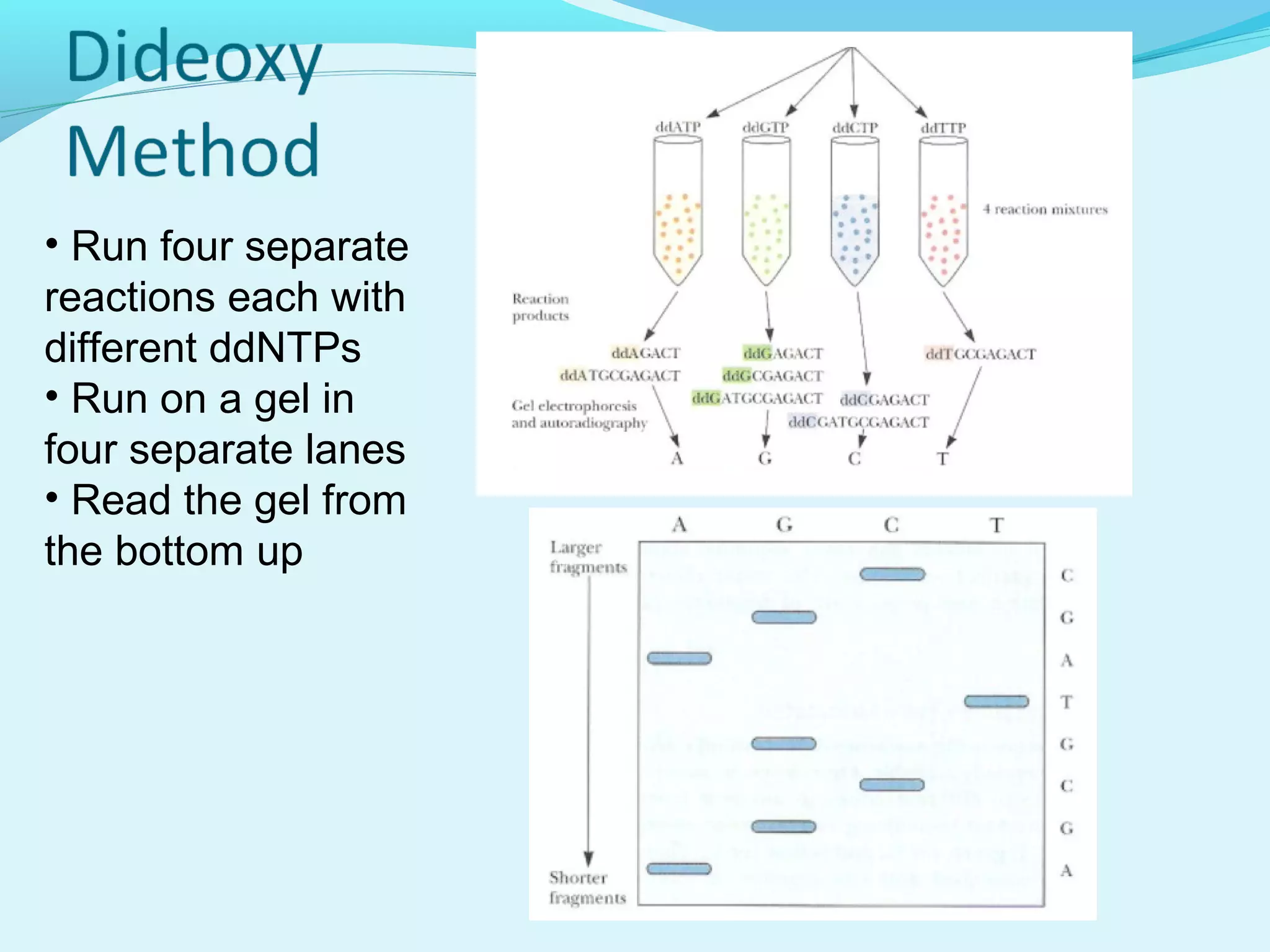
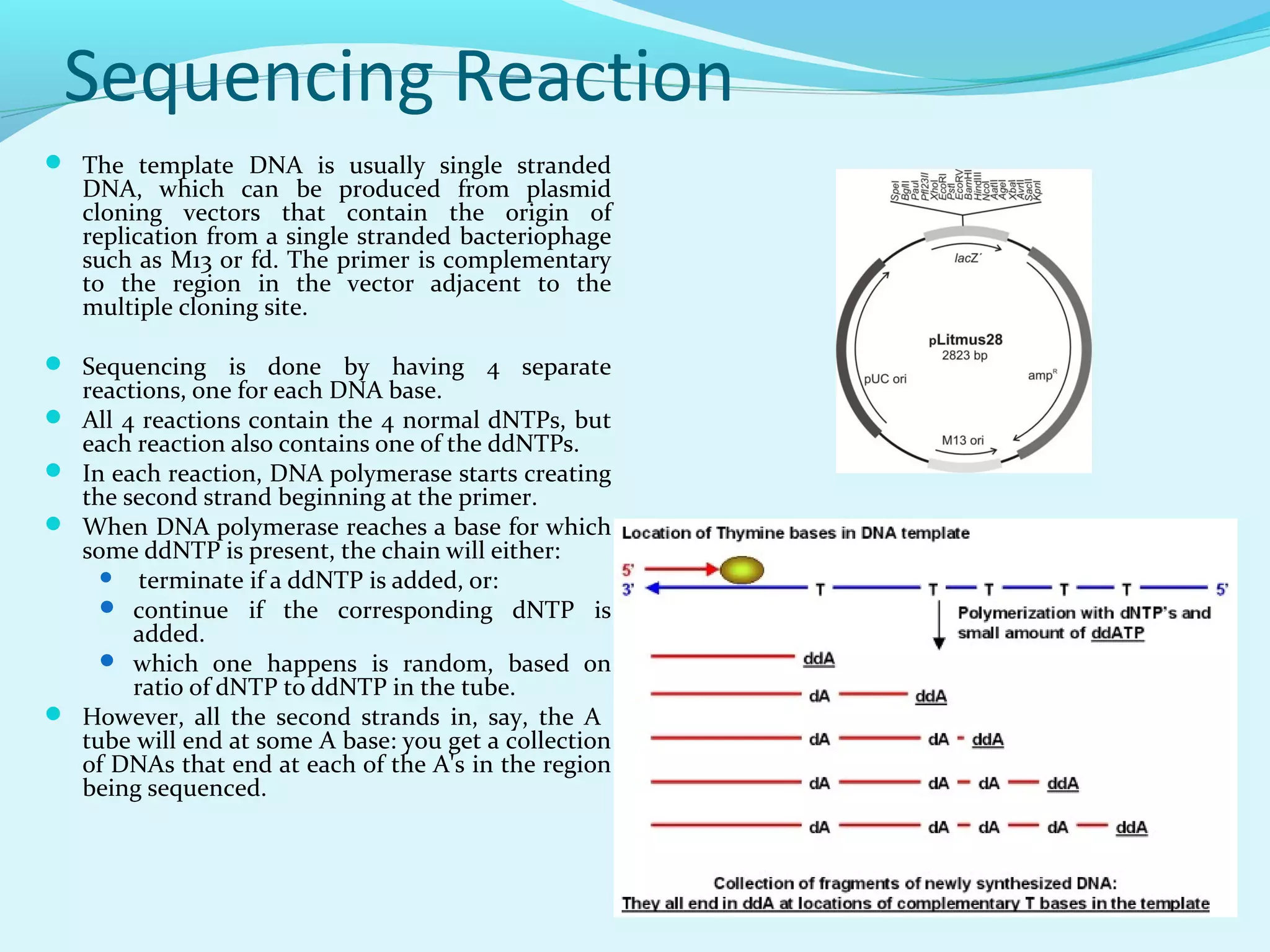
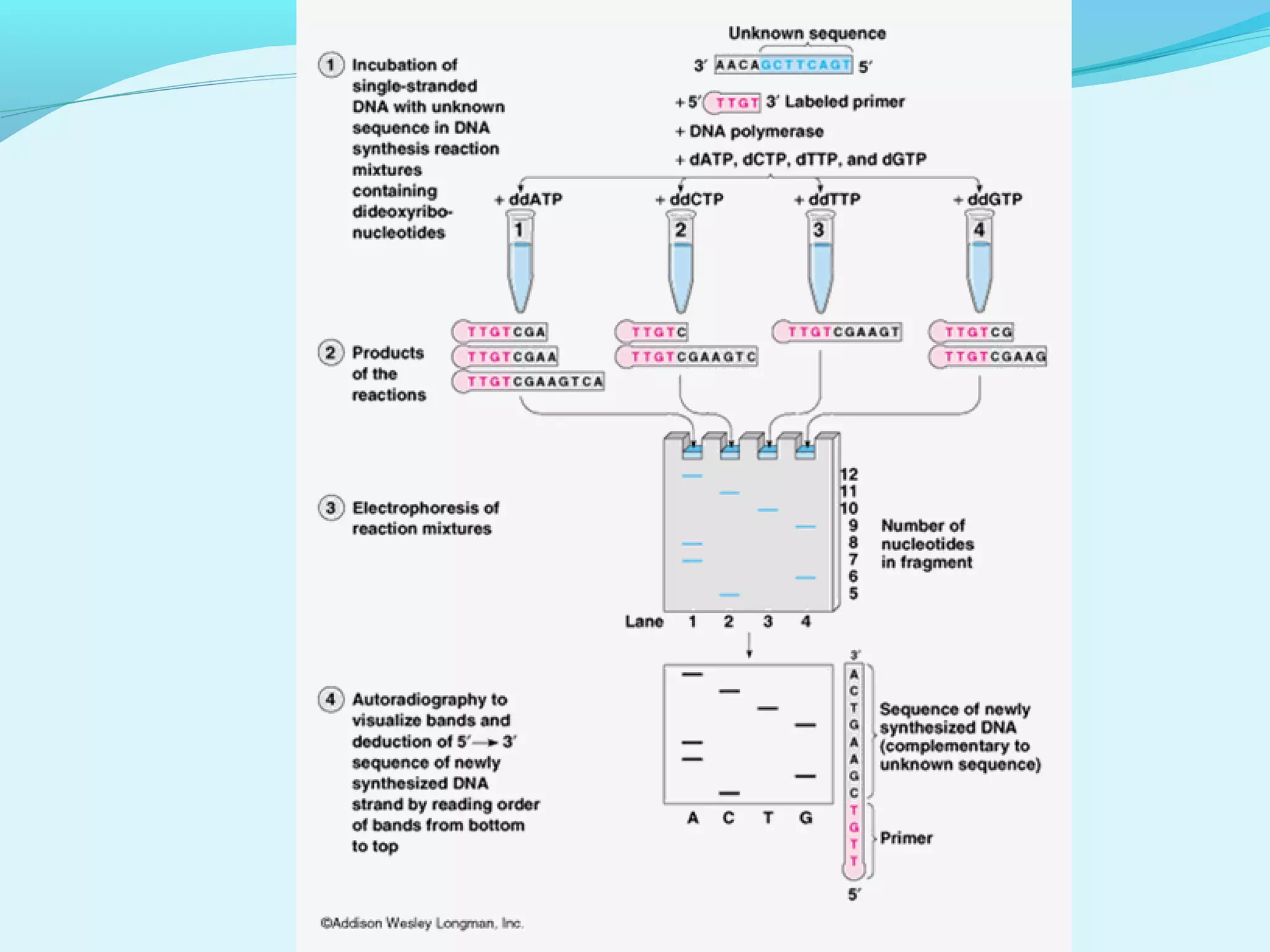
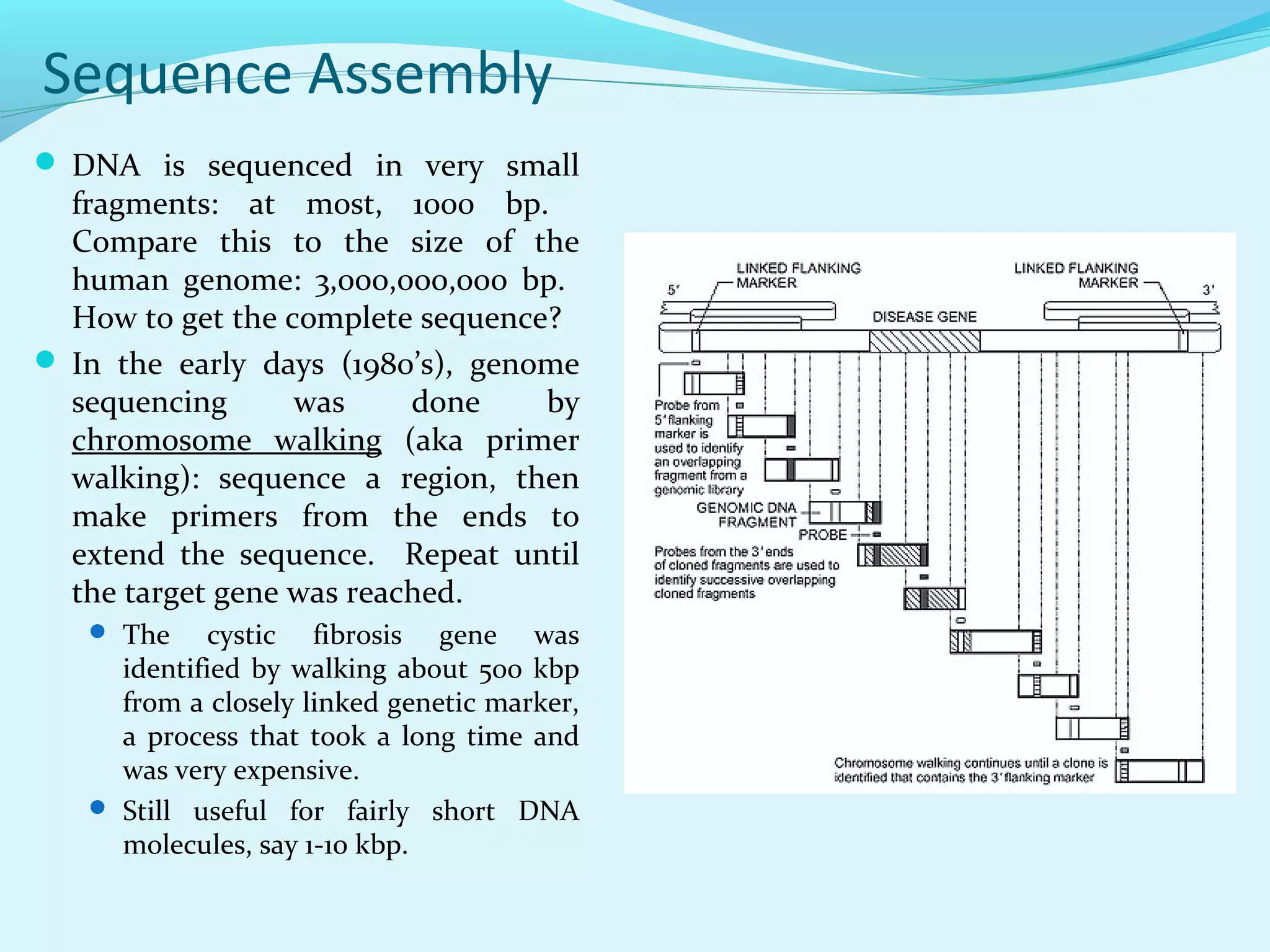
1. DNA sequencing involves determining the order of nucleotide bases in DNA. The original chain termination or dideoxy method developed by Sanger is still widely used for small DNA segments.
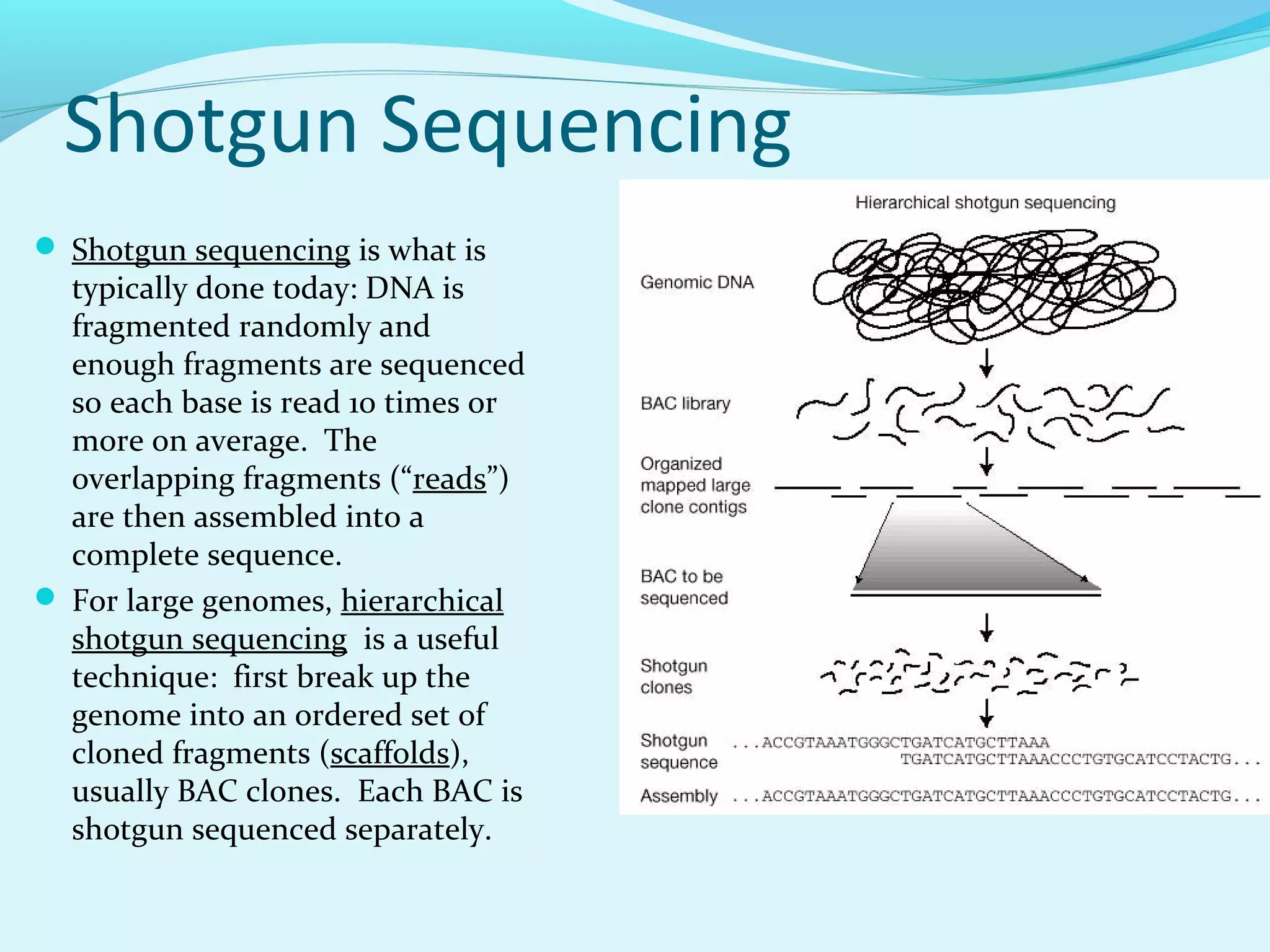
2. Whole genome shotgun sequencing breaks large genomes into fragments that are sequenced and then reassembled, allowing sequencing of entire genomes.
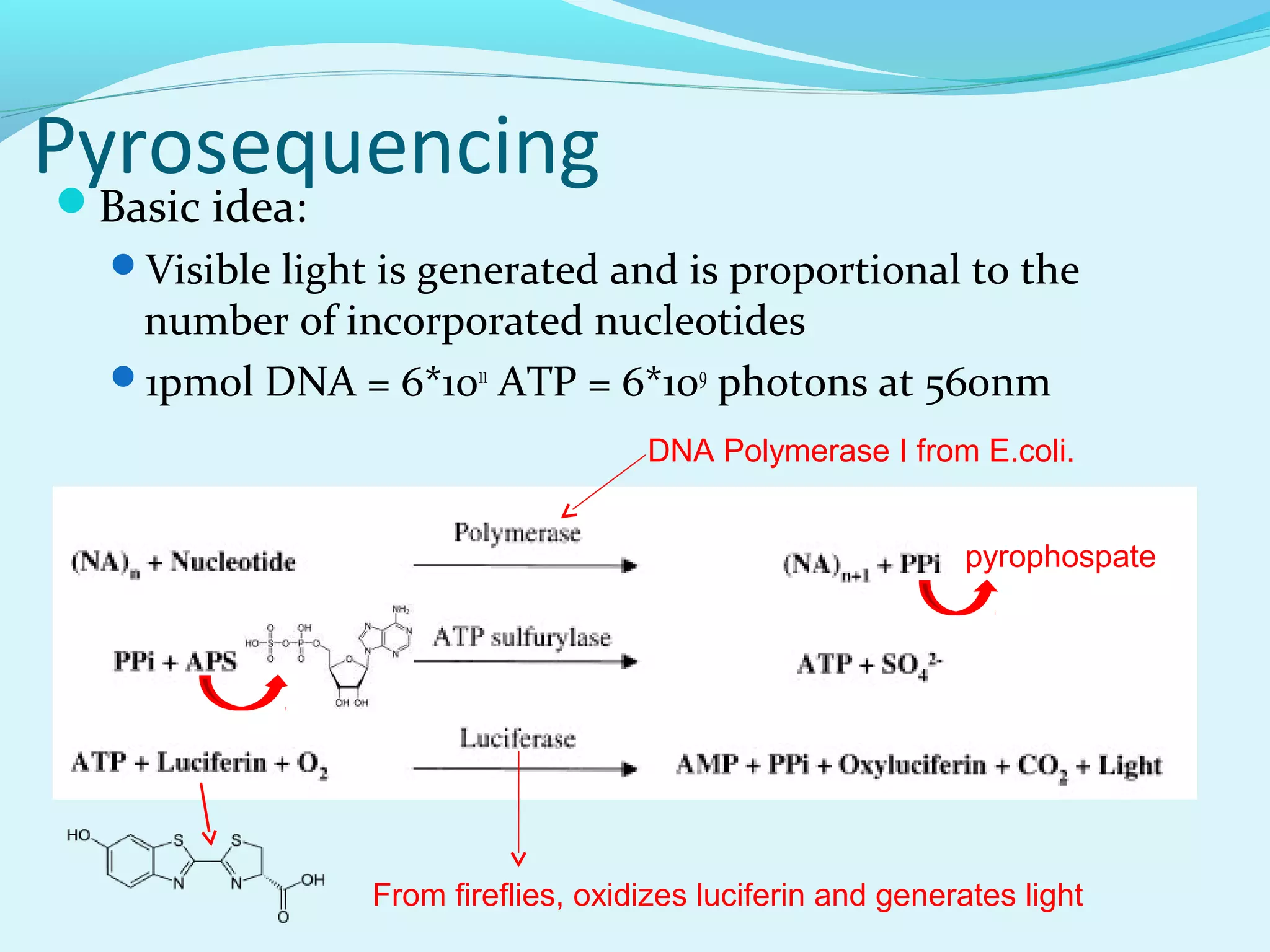
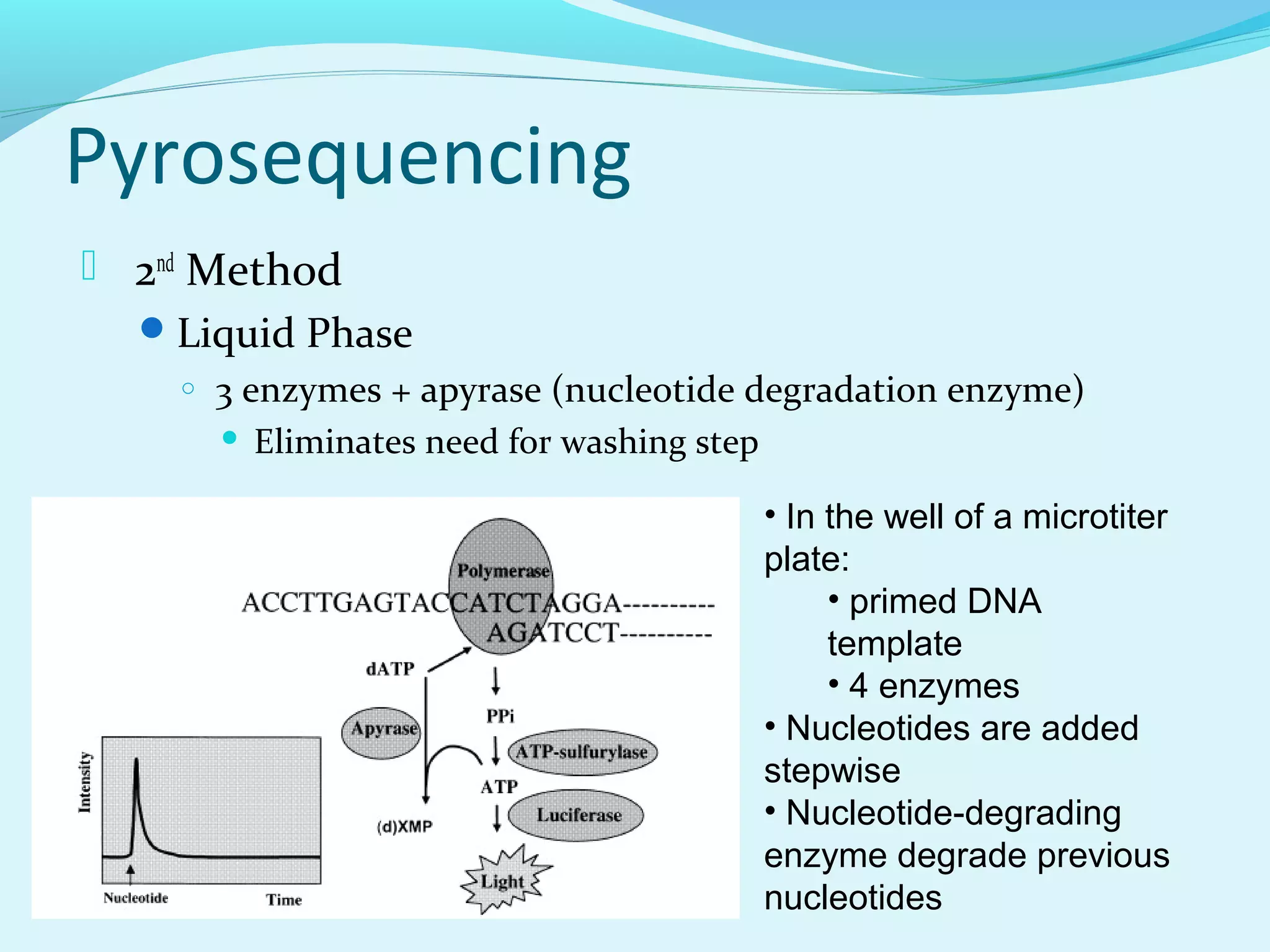
3. Pyrosequencing is a sequencing by synthesis method that uses a bioluminescent reaction to determine nucleotides added, enabling accurate and fast sequencing.