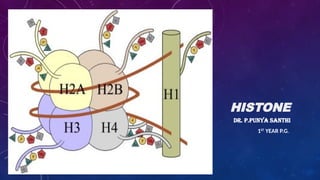
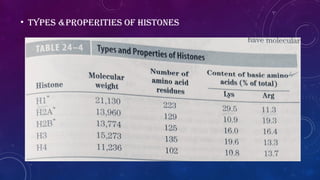
The document provides an overview of histones, which are alkaline proteins critical for DNA organization in eukaryotic cells and play roles in gene regulation, DNA replication, and repair. It discusses different types of histones, their modifications such as acetylation and methylation, and introduces the concept of the 'histone code,' which links these modifications to gene expression. Additionally, it mentions diseases associated with histone modifications and techniques for studying these modifications.