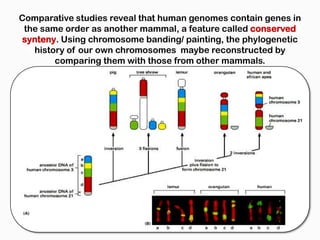
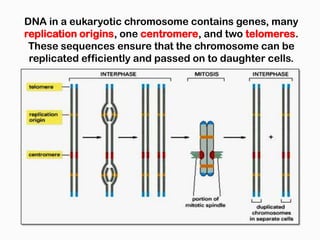
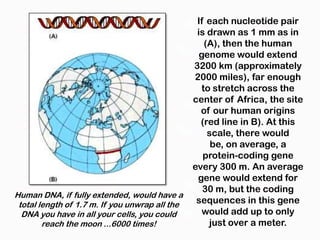
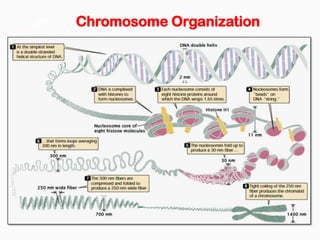
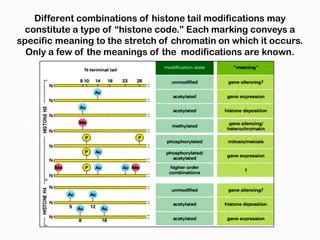
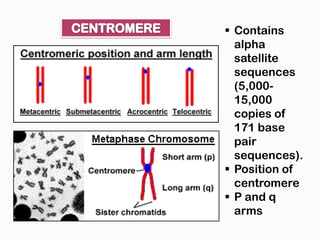
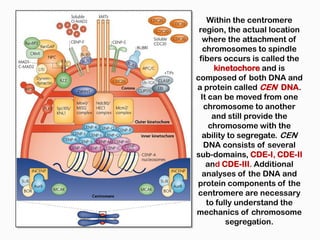
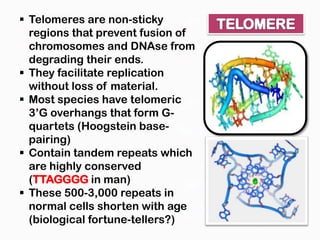
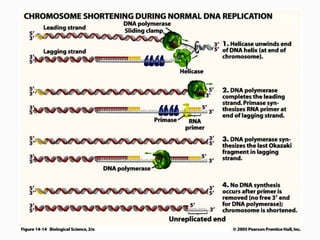
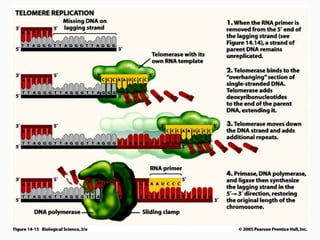
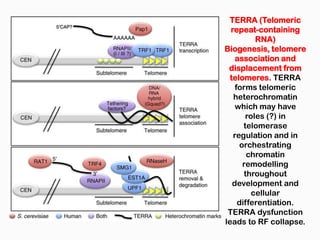
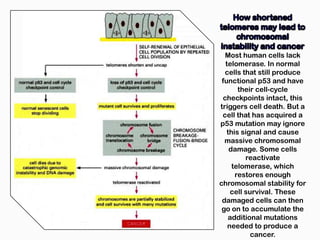
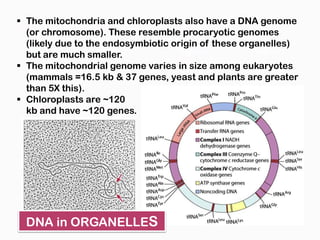
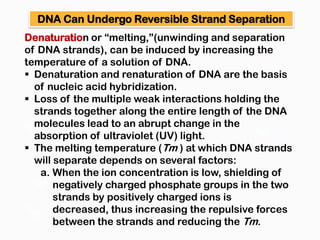
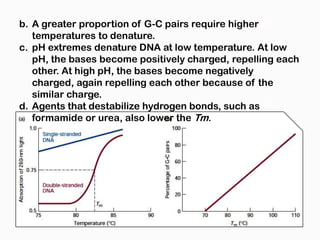
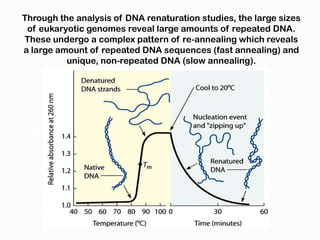
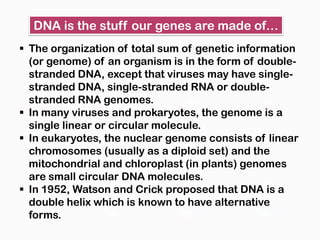
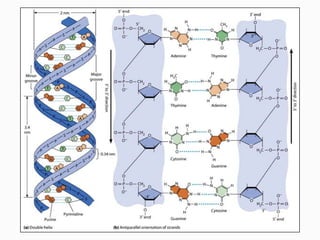
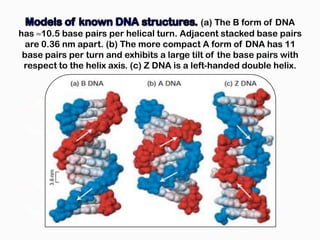
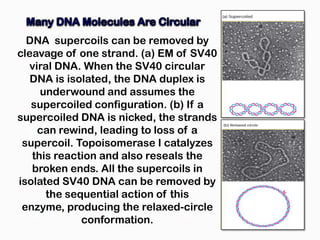
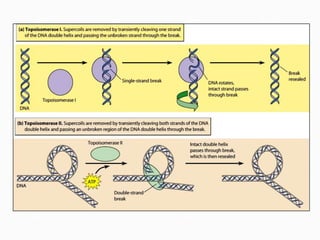
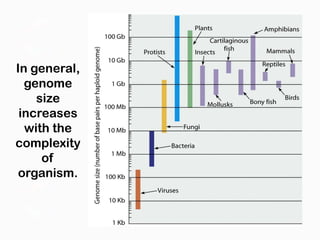
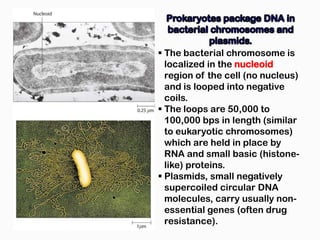
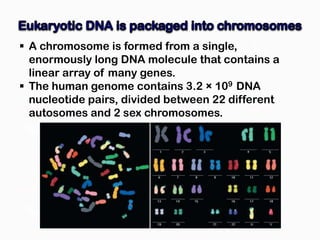
DNA is the genetic material found in the nucleus of eukaryotic cells and in the chromosomes of prokaryotes. It exists in several forms, including linear chromosomes in eukaryotes and circular chromosomes in prokaryotes and organelles. DNA is made up of a double helix structure stabilized by hydrogen bonding between complementary nucleotide base pairs. The structure of DNA allows it to efficiently store and transmit genetic information.


![Chromosomal banding patterns and multicolor FISH are
used to analyze human anomalies
Characteristic chromosomal translocations are associated with
certain genetic disorders and specific types of cancers. In nearly
all patients with chronic myelogenous leukemia, the leukemic
cells contain the Philadelphia chromosome, a shortened
chromosome 22 [der (22)], and an abnormally long chromosome
9 [der (9)]. These result from a translocation between normal
chromosomes 9 and 22.](https://image.slidesharecdn.com/dnaandchromosomes-111109071547-phpapp02/85/Dna-and-chromosomes-11-320.jpg)