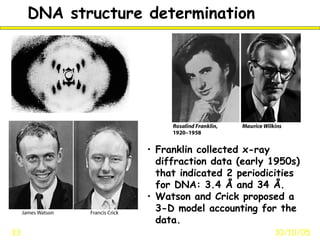
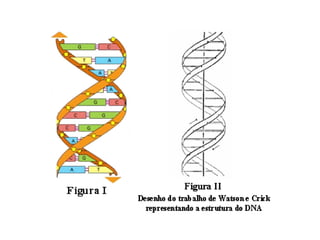
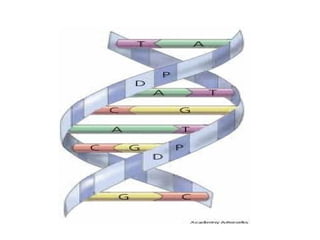
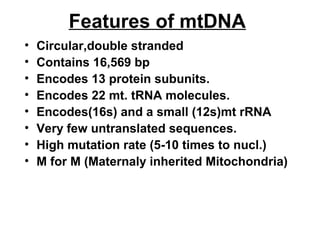
Franklin collected x-ray diffraction data in the early 1950s that showed DNA has two periodicities: 3.4 Å and 34 Å. Watson and Crick then proposed a 3D model of DNA that accounted for Franklin's data, representing the first model of the DNA double helix structure. This established that DNA is made of two antiparallel strands coiled around each other.