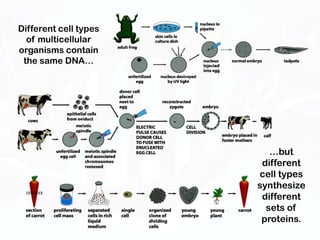
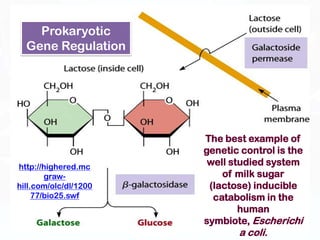
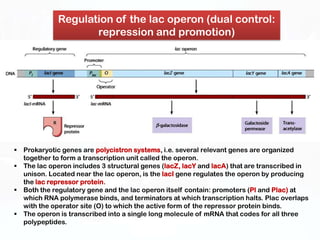
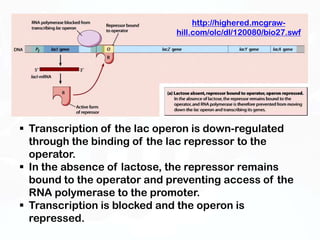
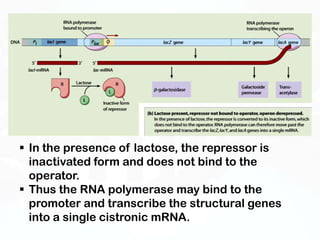
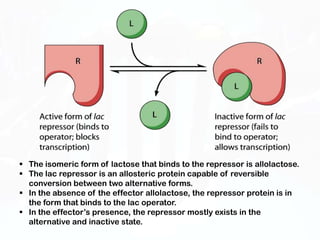
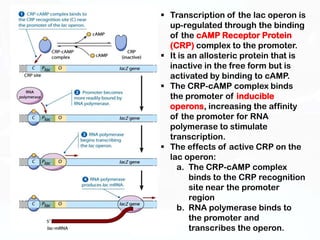
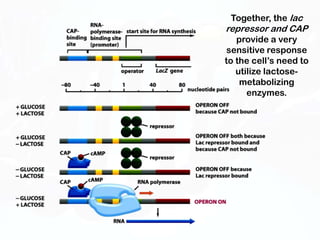
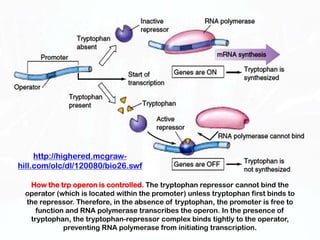
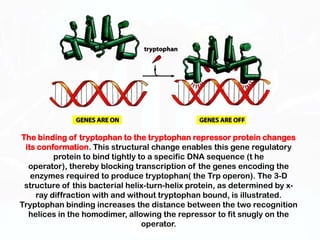
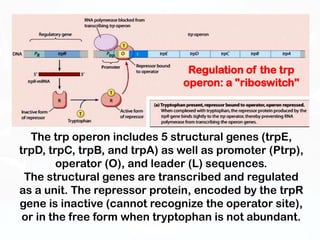
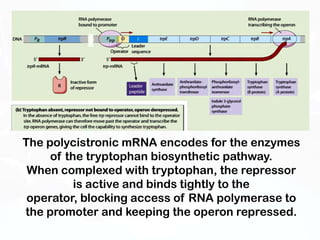
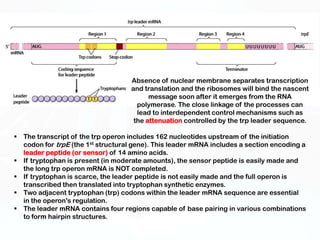
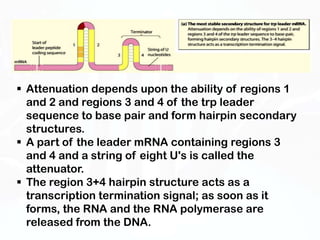
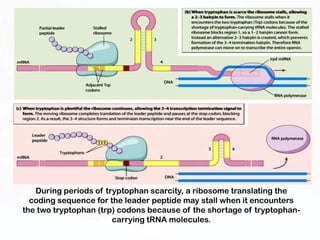
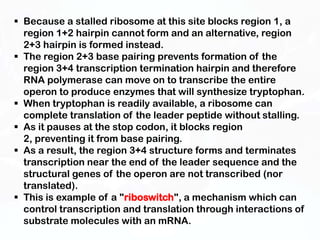
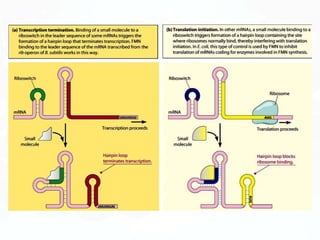
Different cell types contain the same DNA but express different genes and proteins. Gene expression can be regulated at many steps from DNA to RNA to protein. Prokaryotes regulate gene expression through operons, where genes are organized together and transcribed as a single unit. The lac and trp operons are regulated by repressor proteins that bind to operator sites on the DNA and block transcription in the presence of effector molecules like lactose or tryptophan. This document discusses the mechanisms of repression and induction of the lac and trp operons through repressor proteins and RNA polymerase binding.