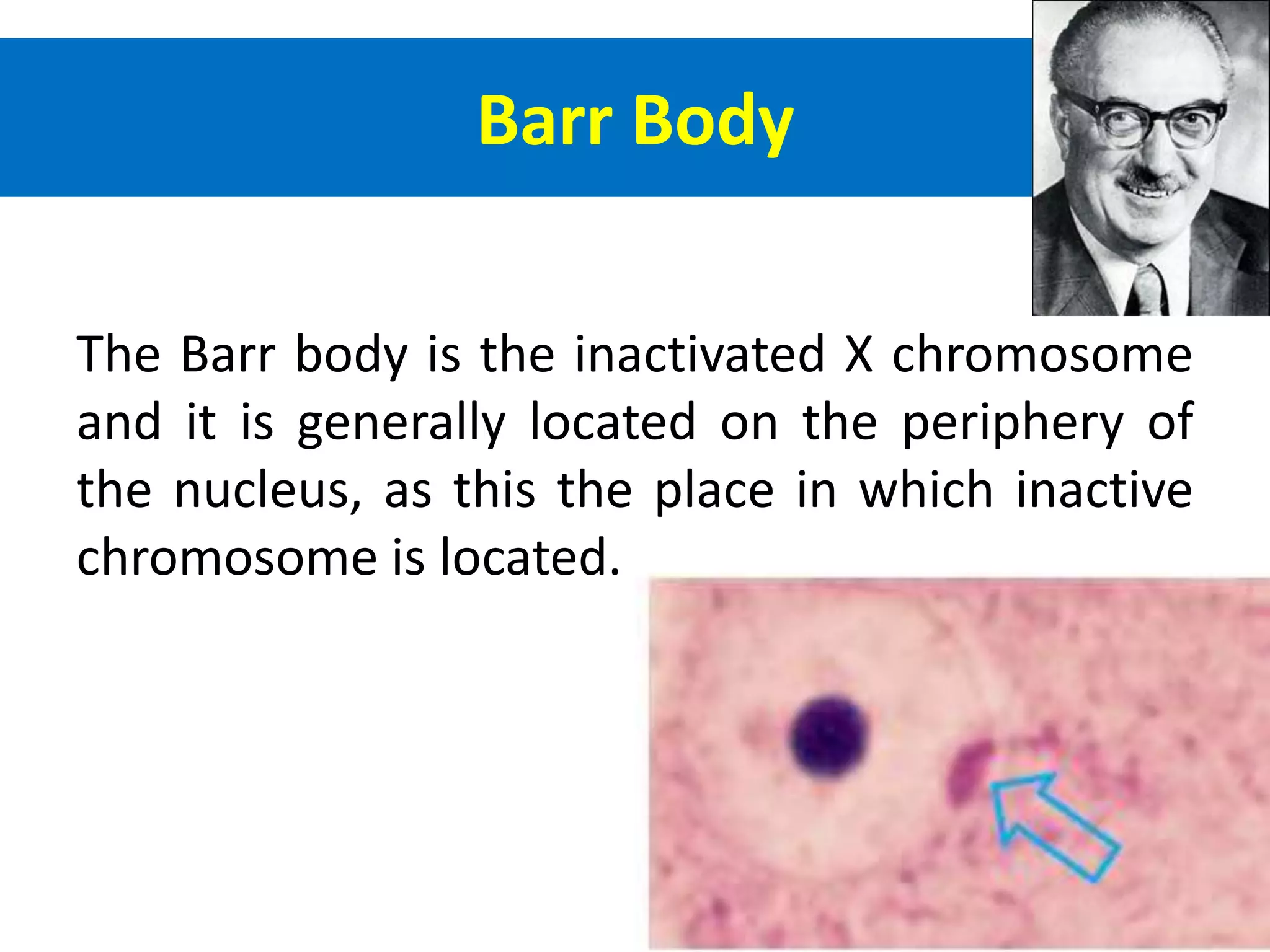
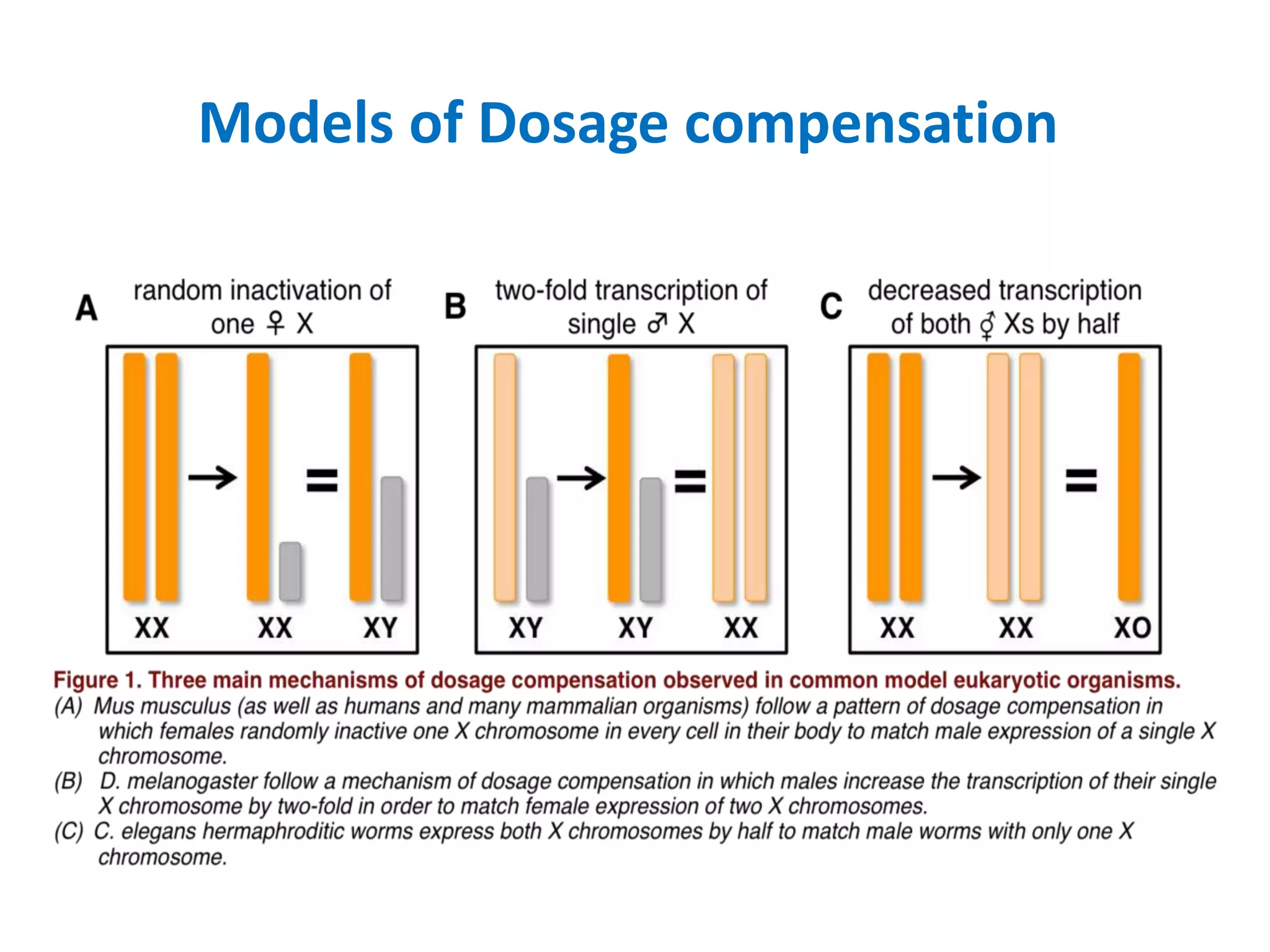
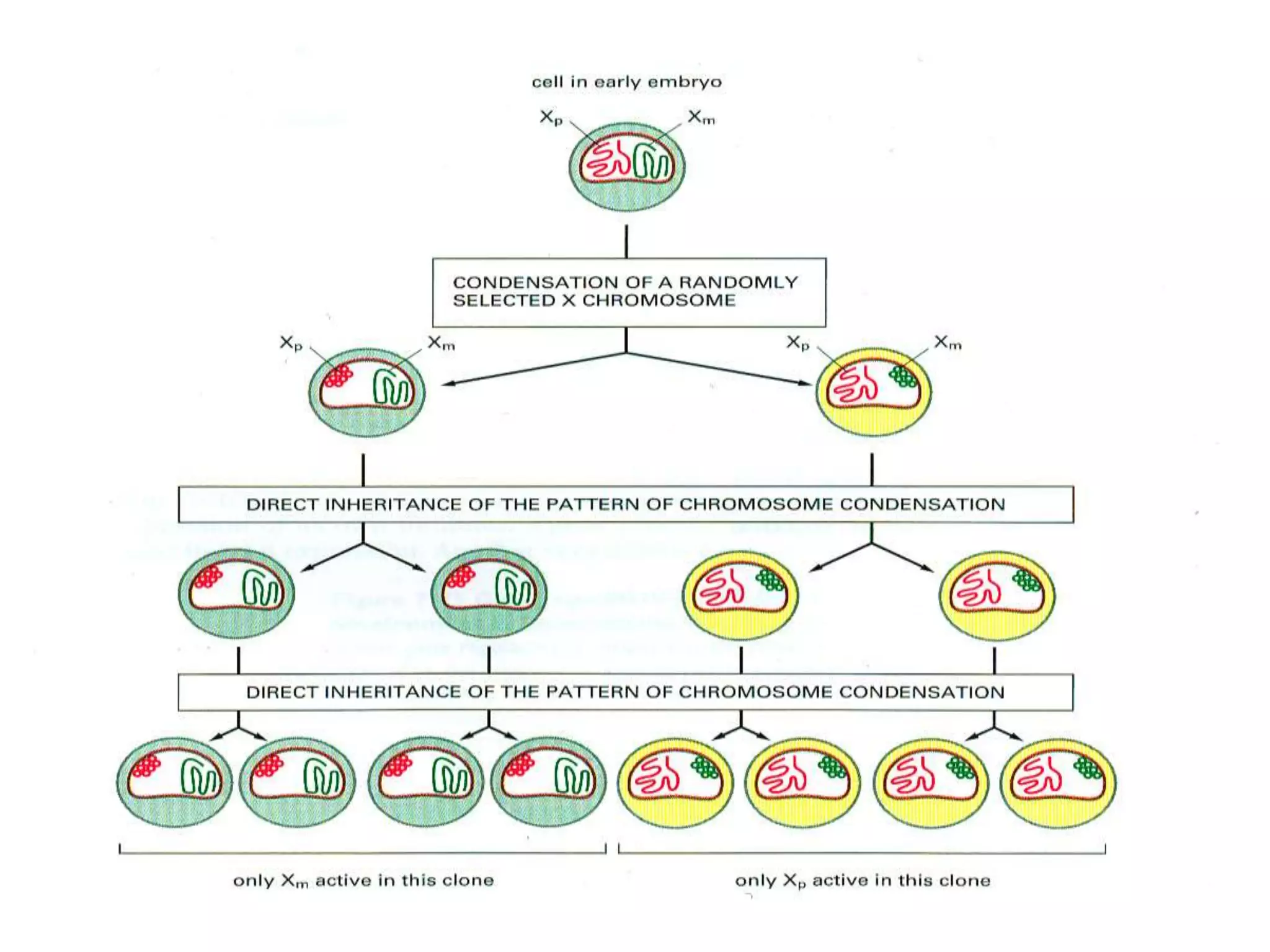
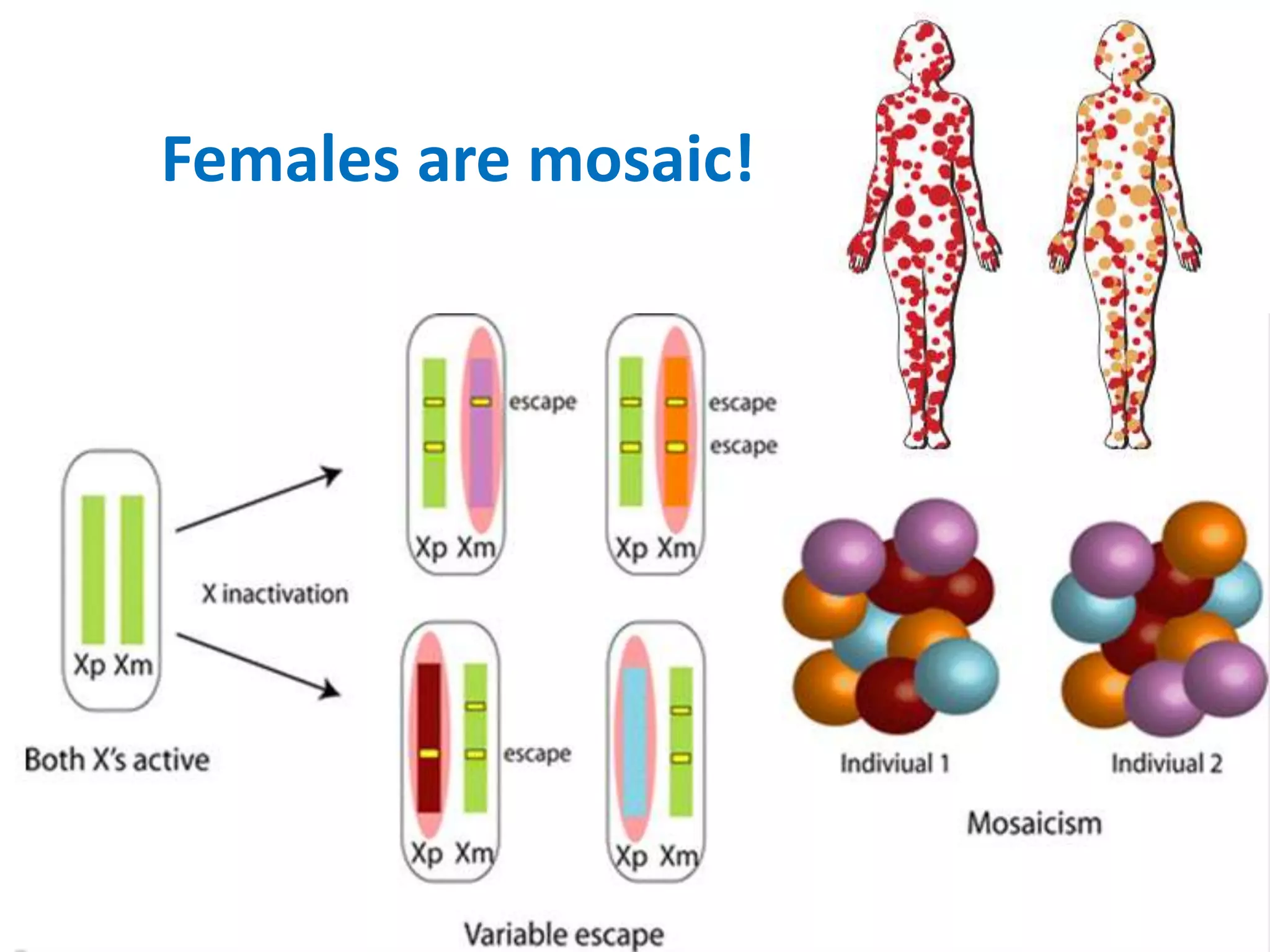
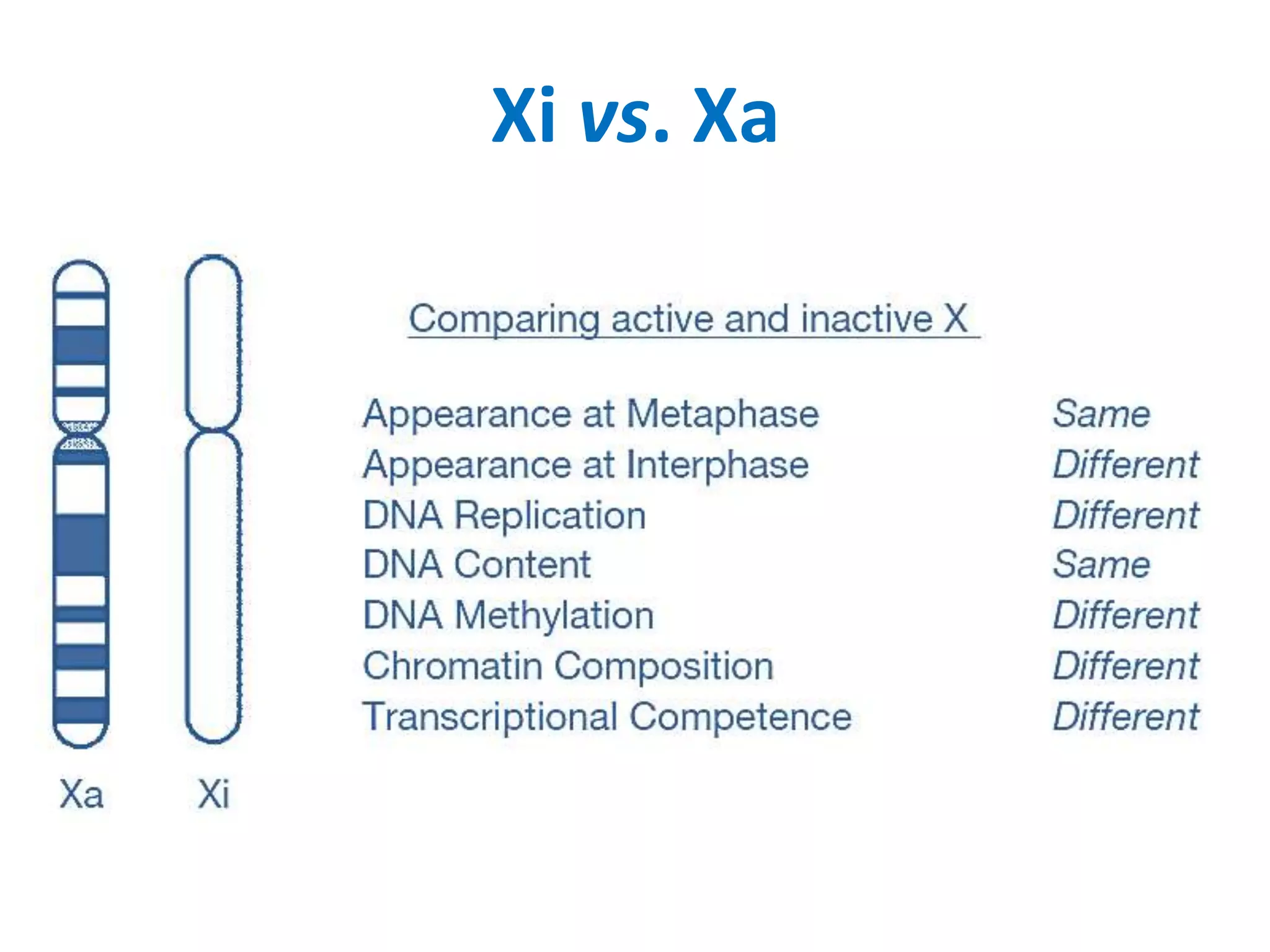
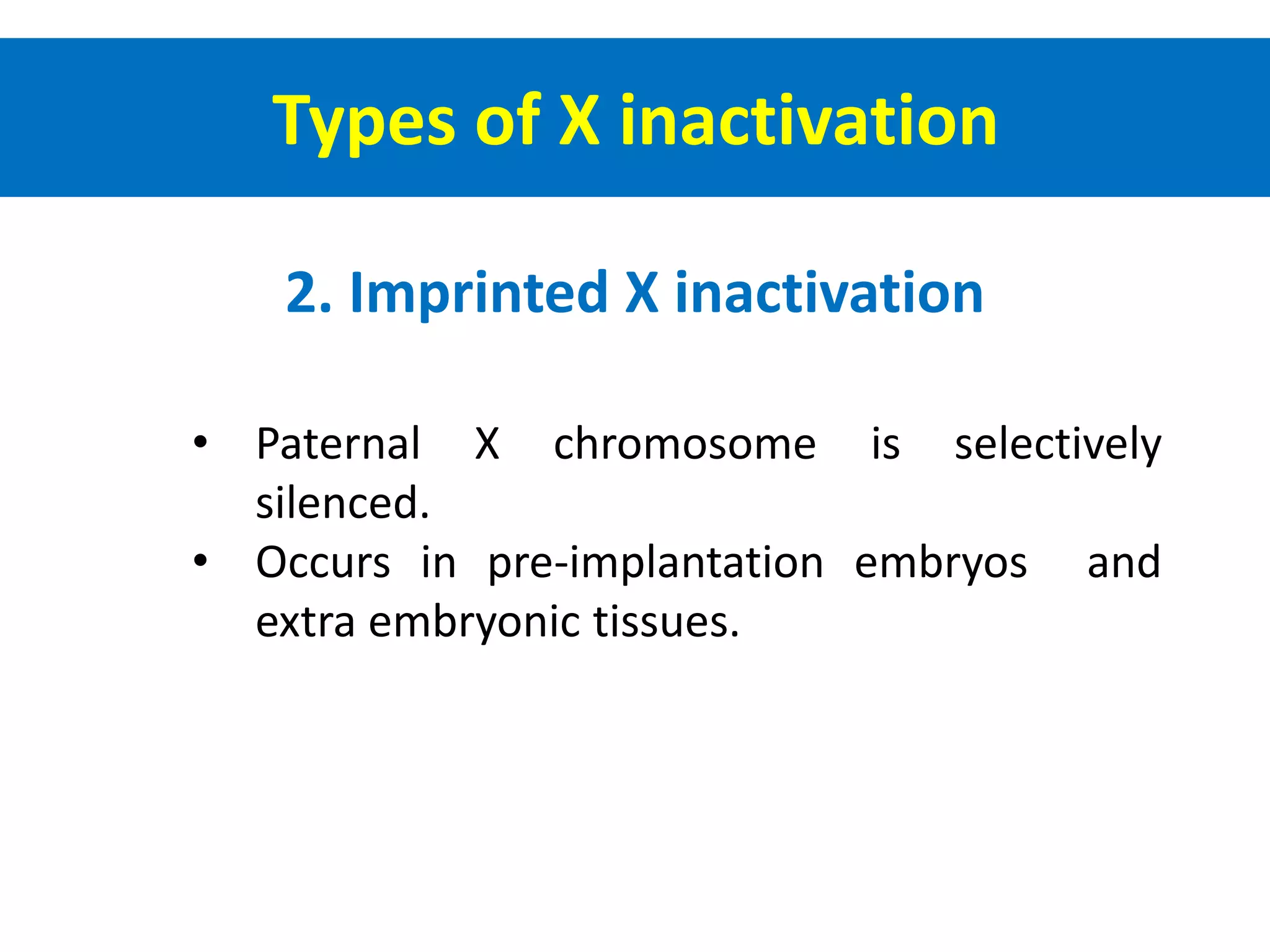
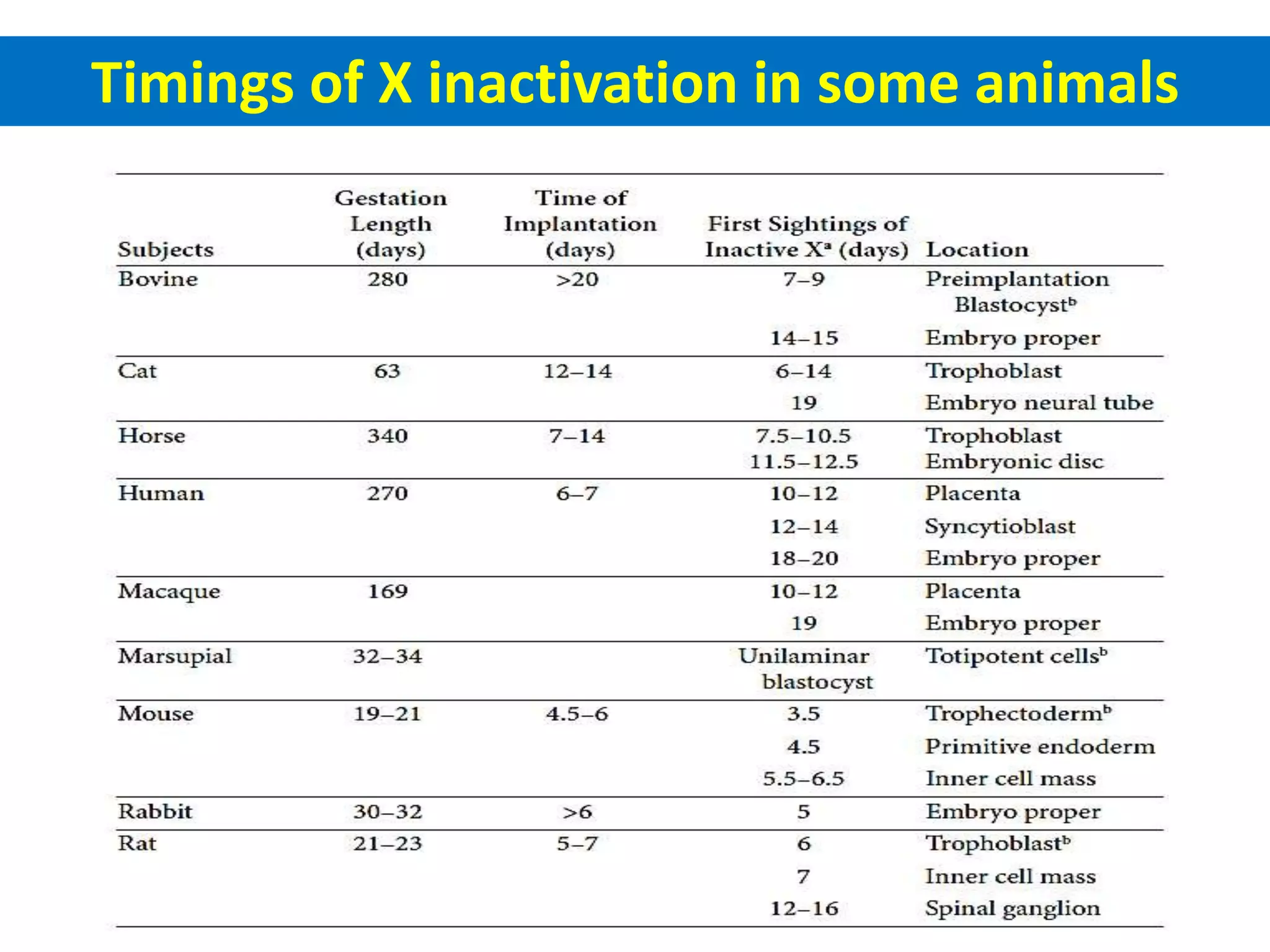
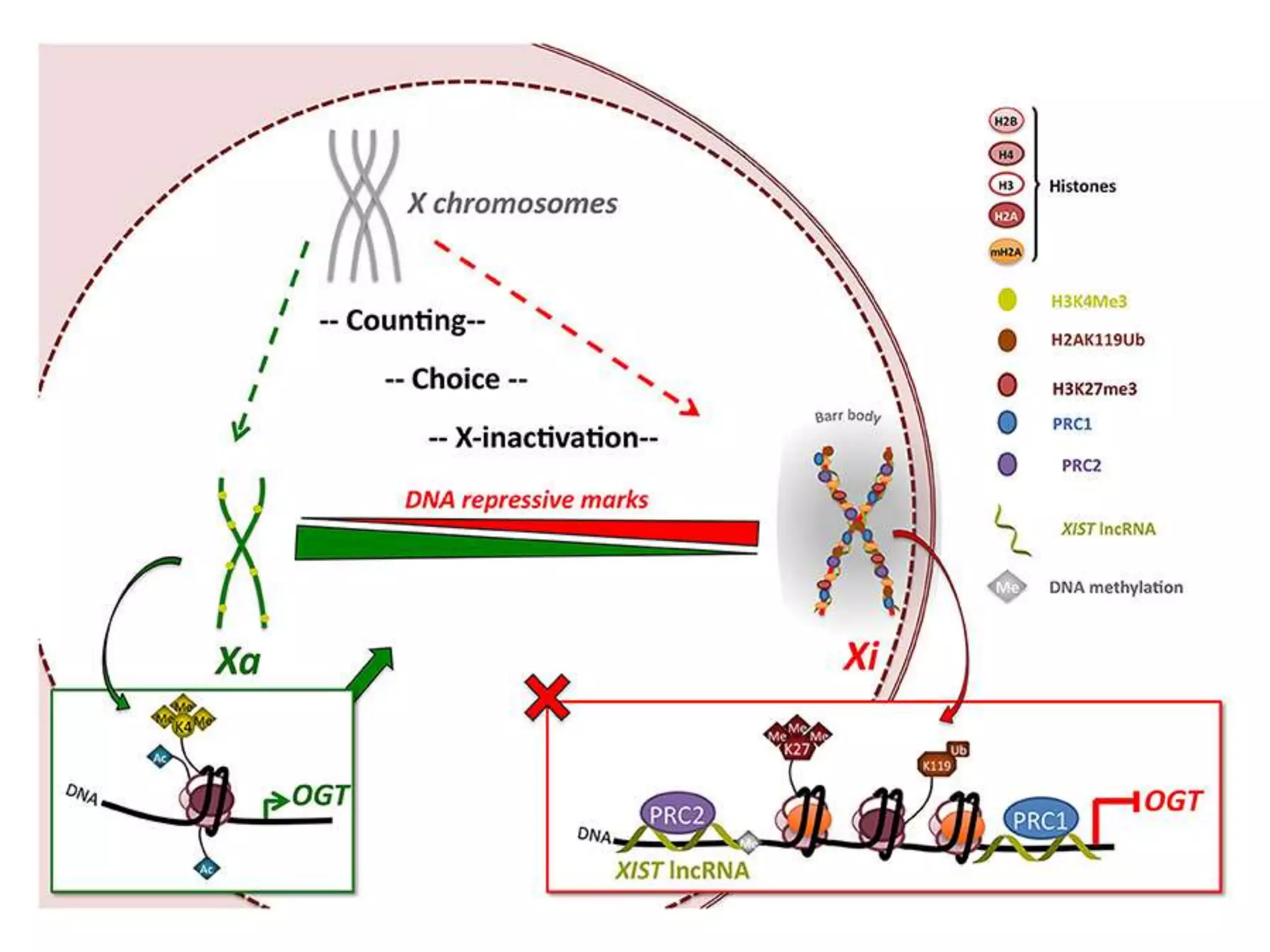
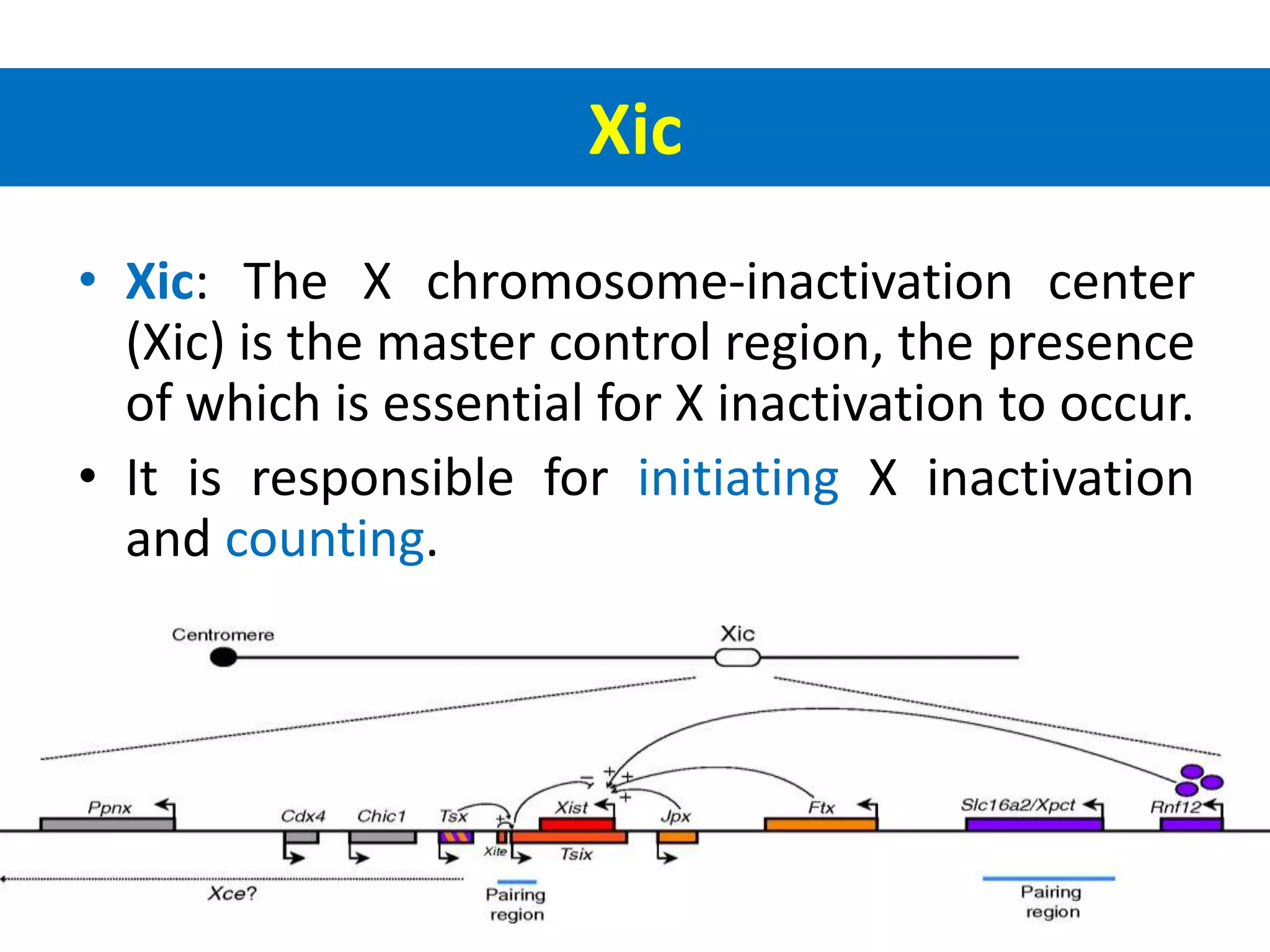
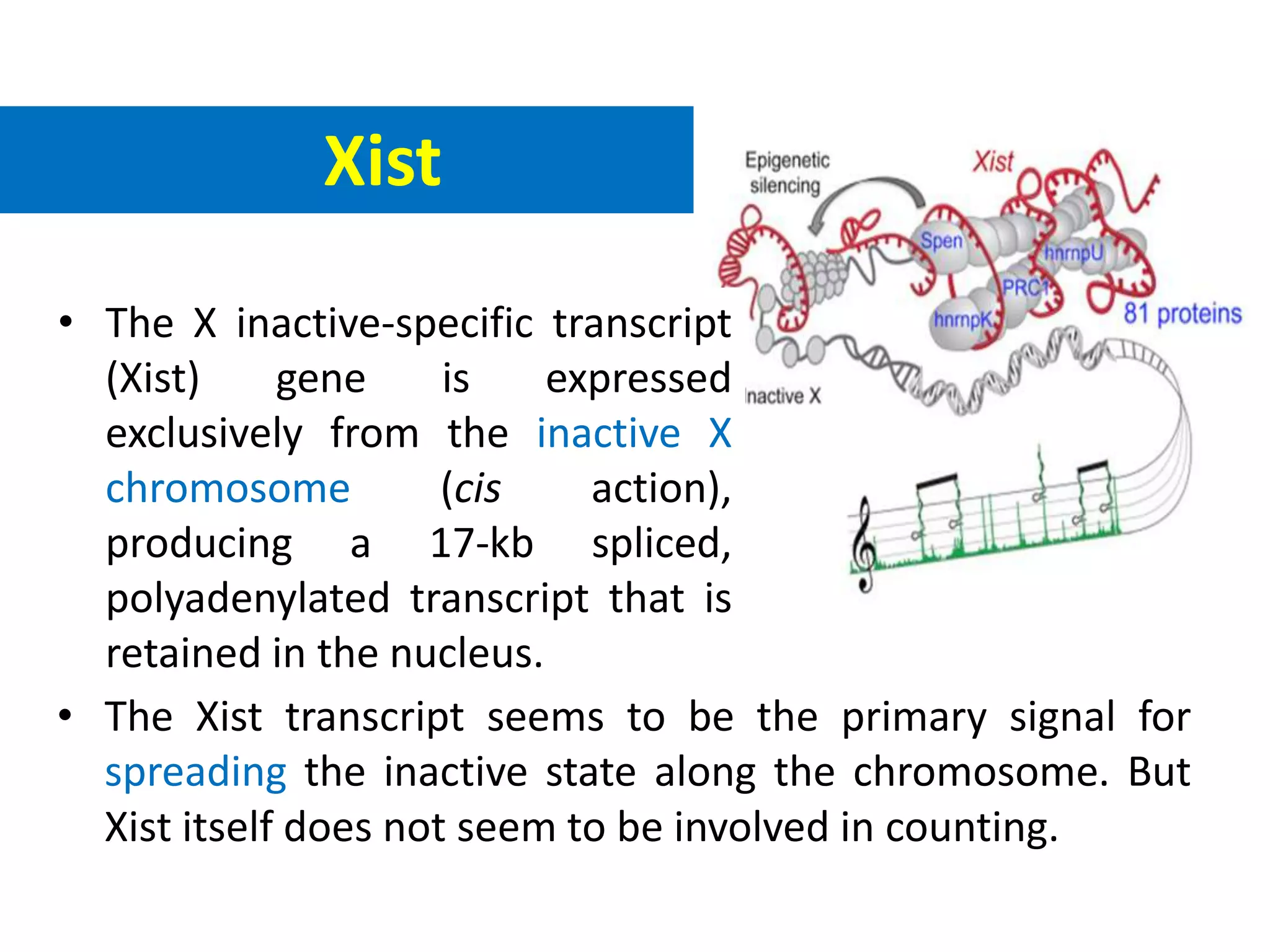
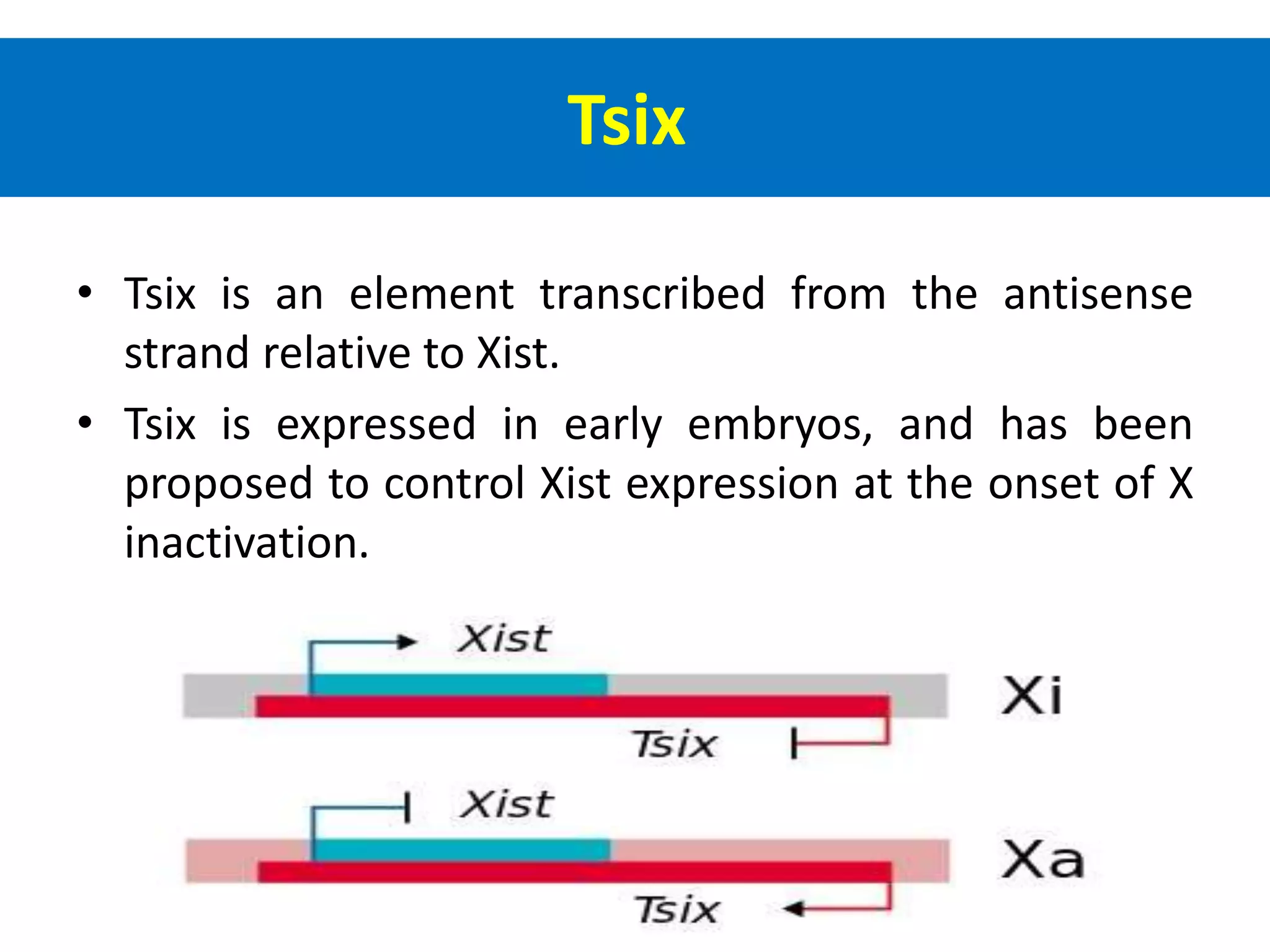
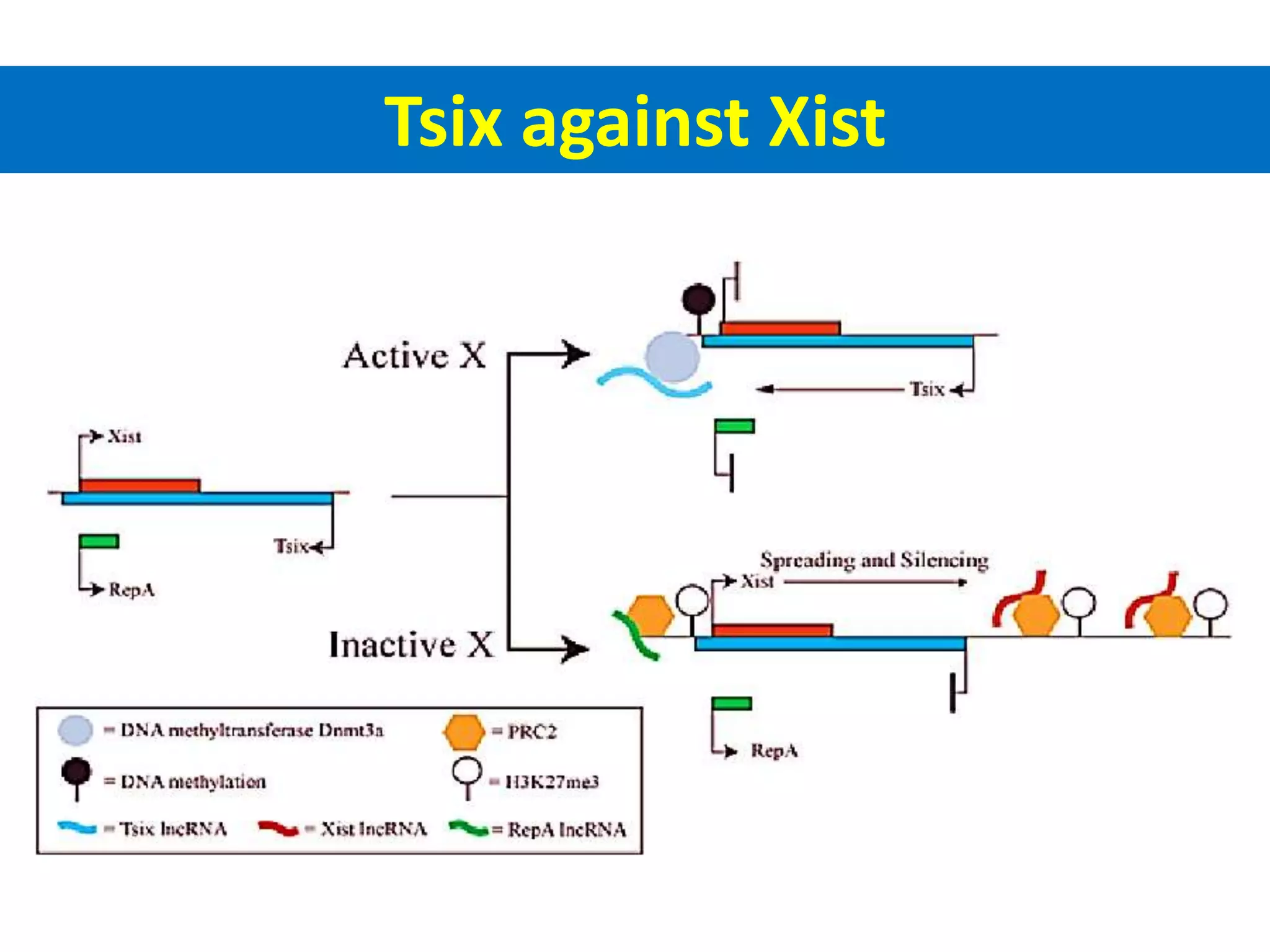
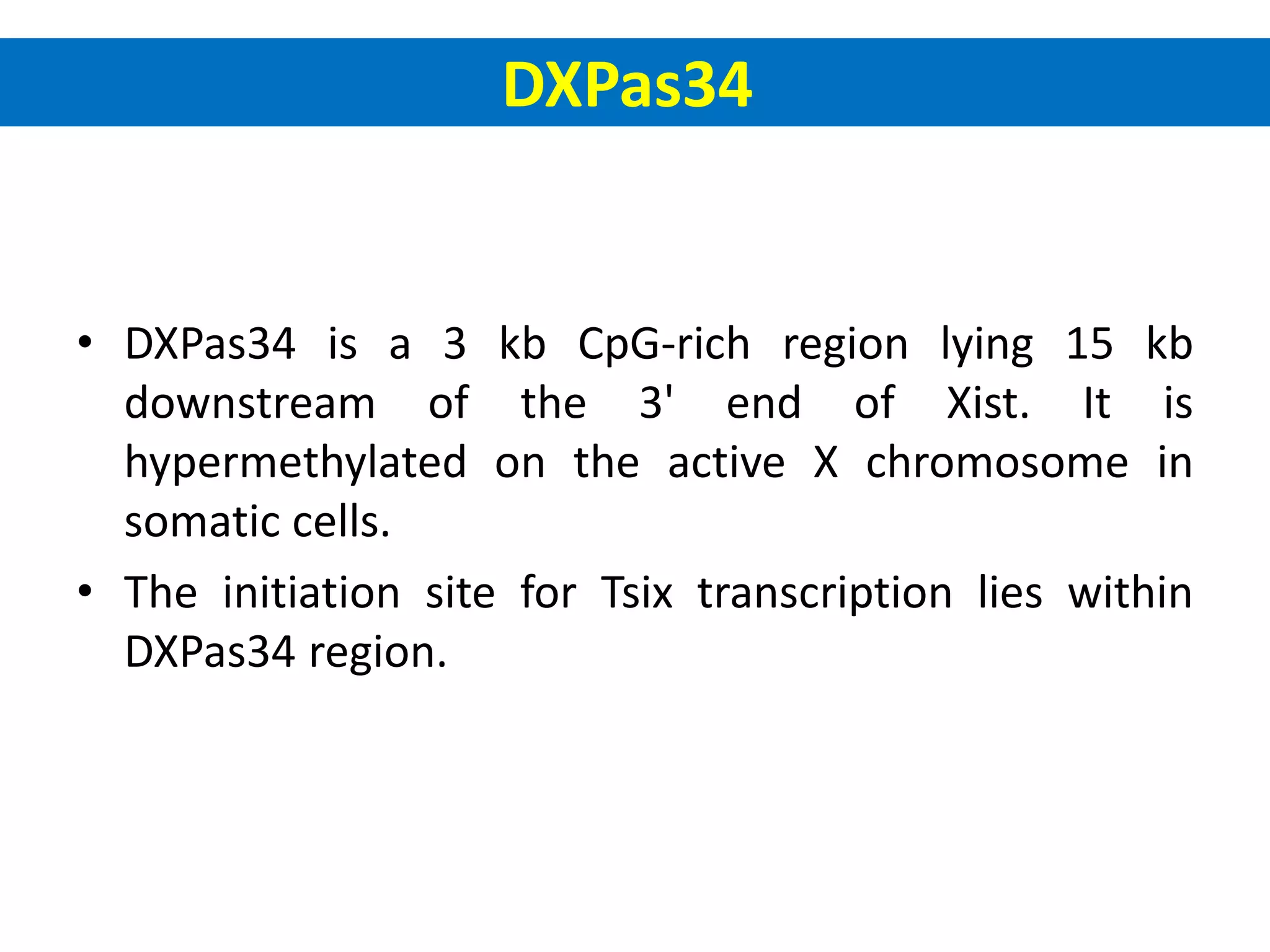
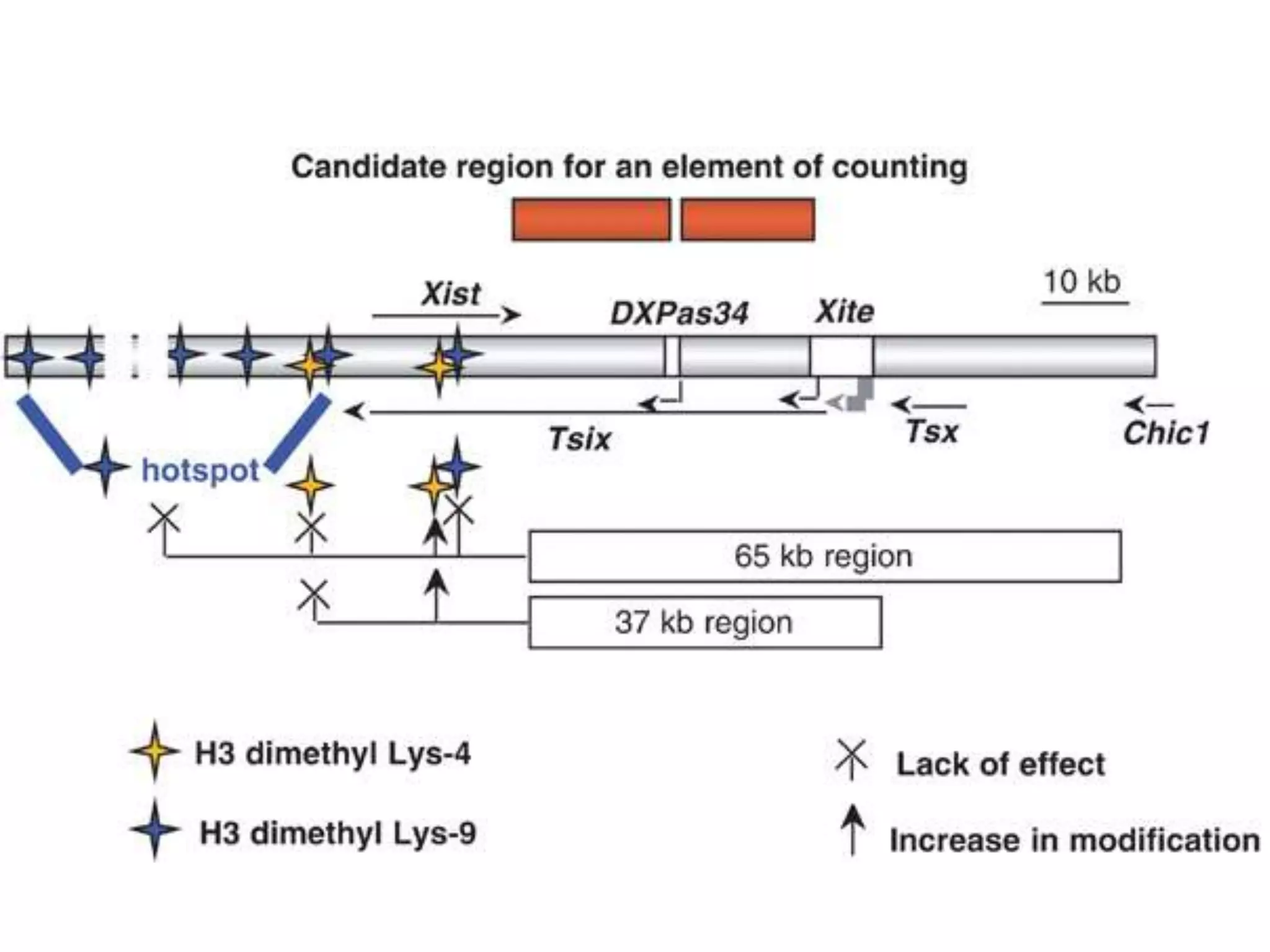
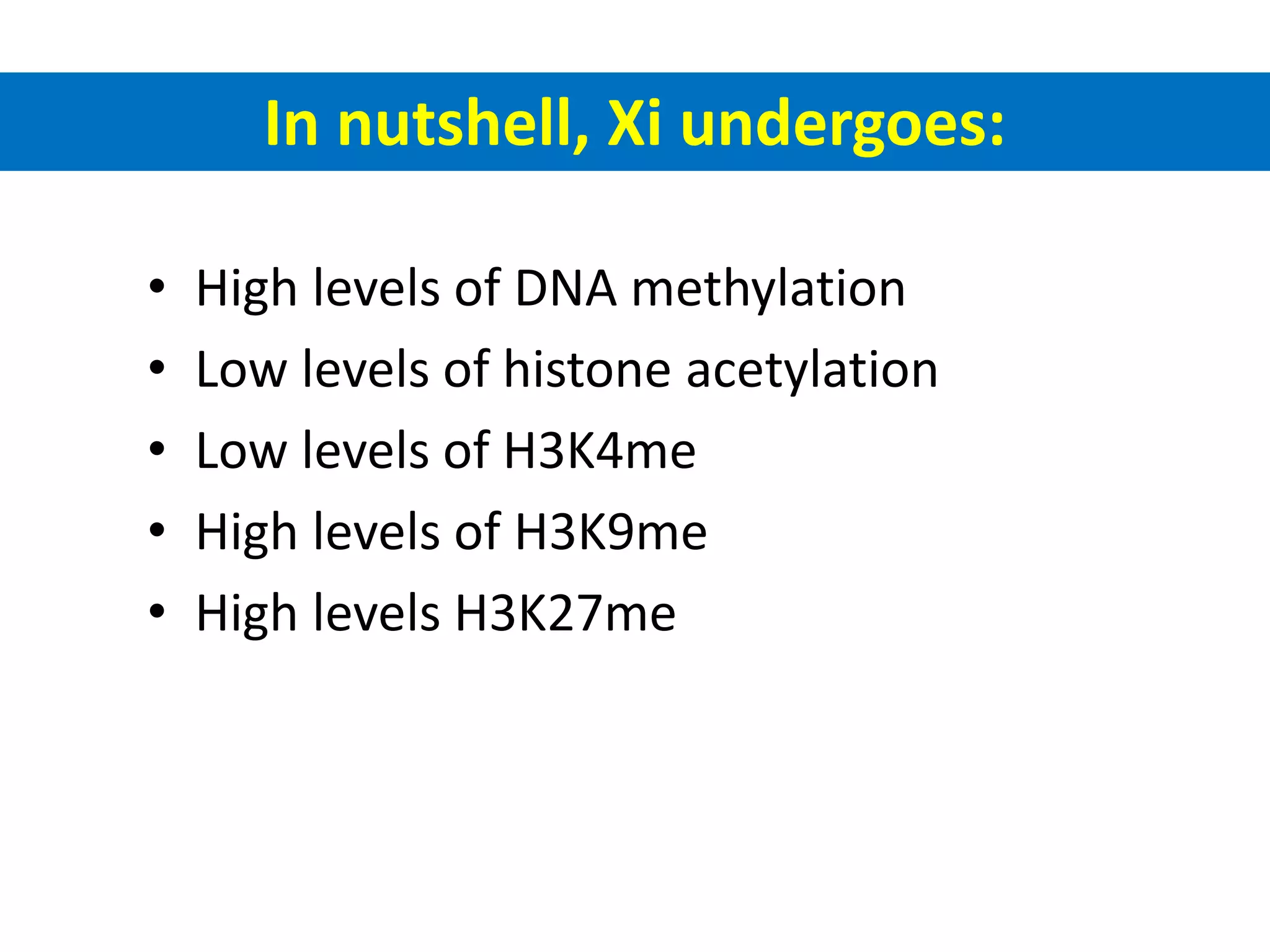
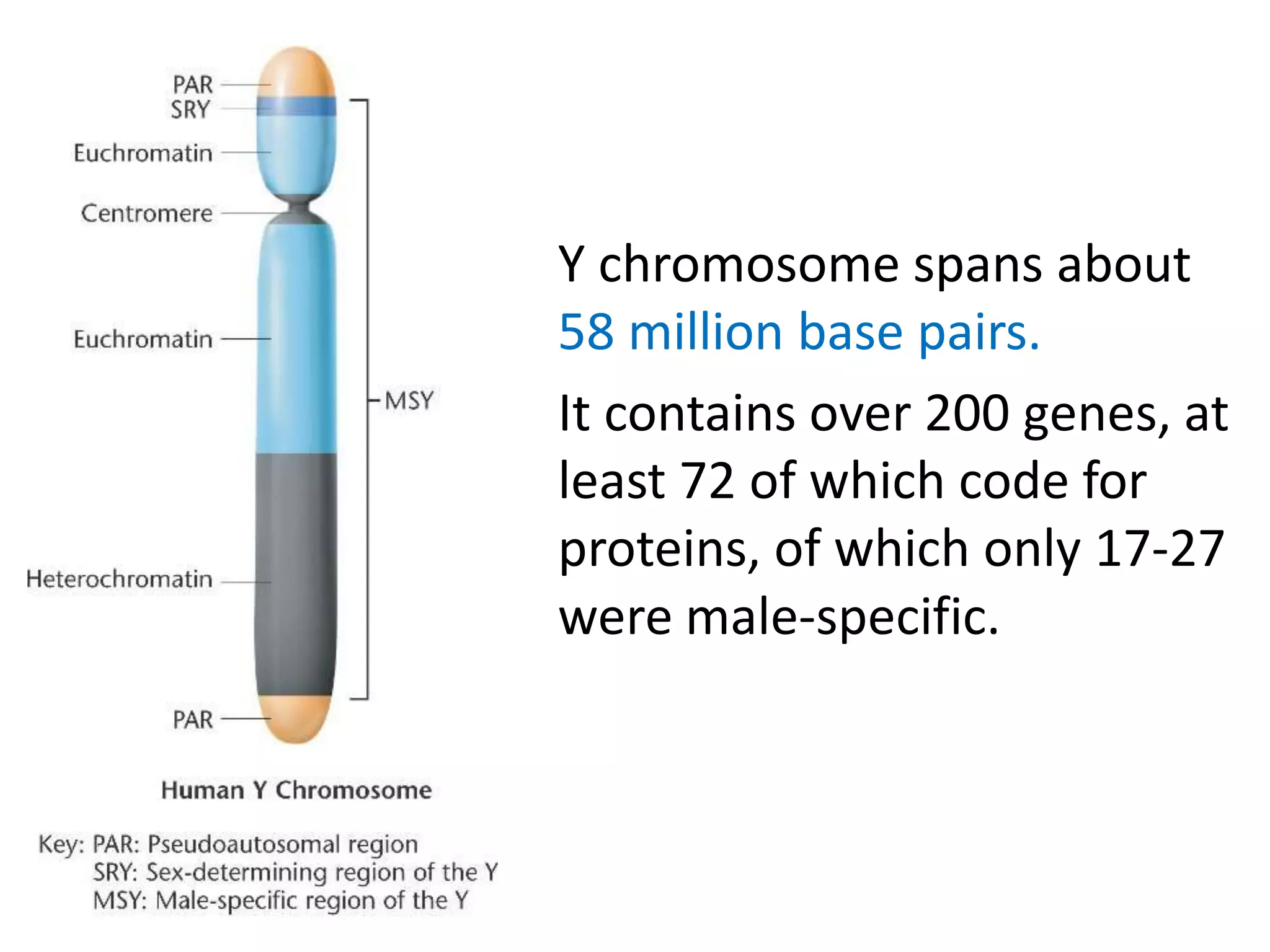
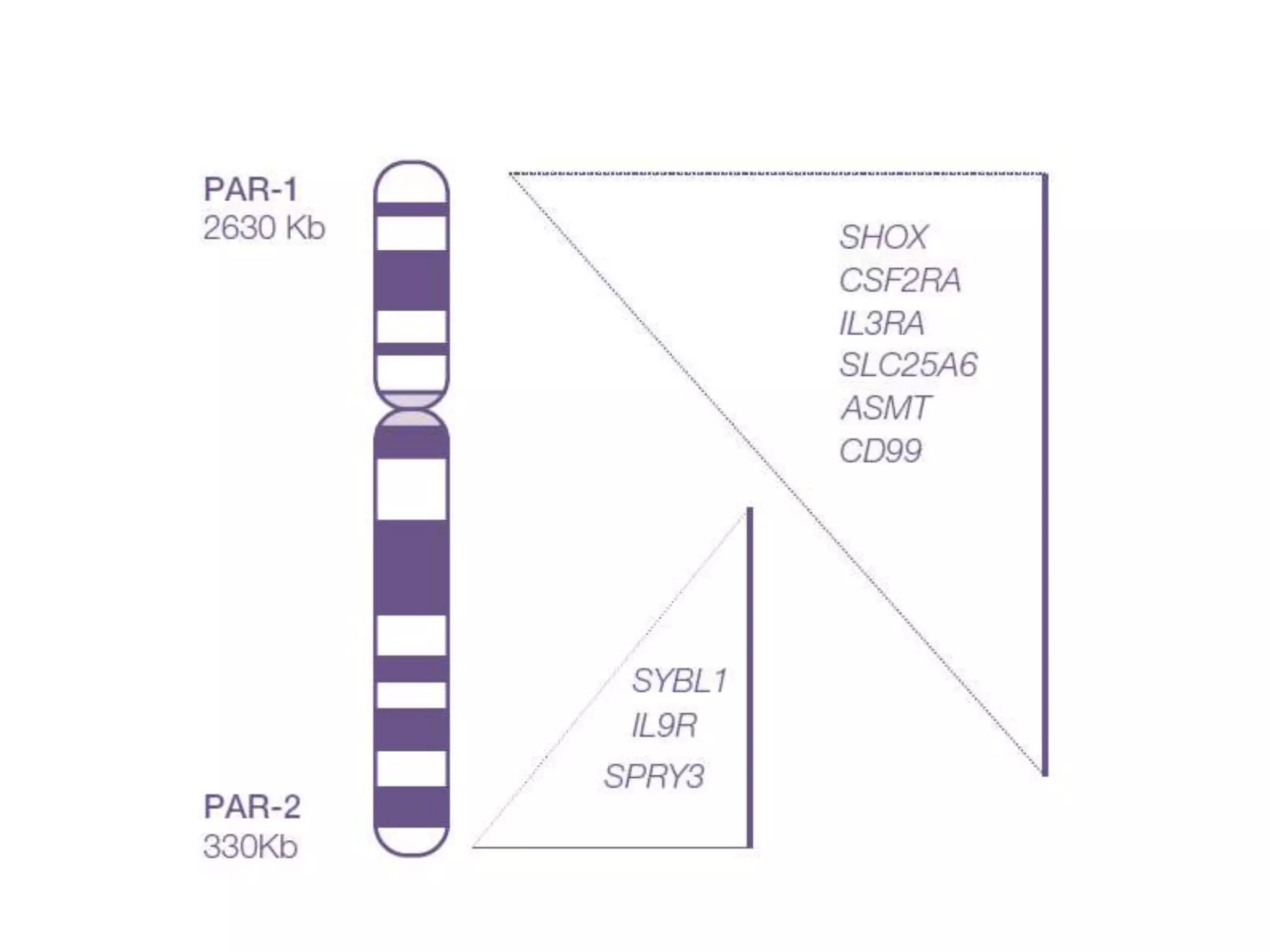
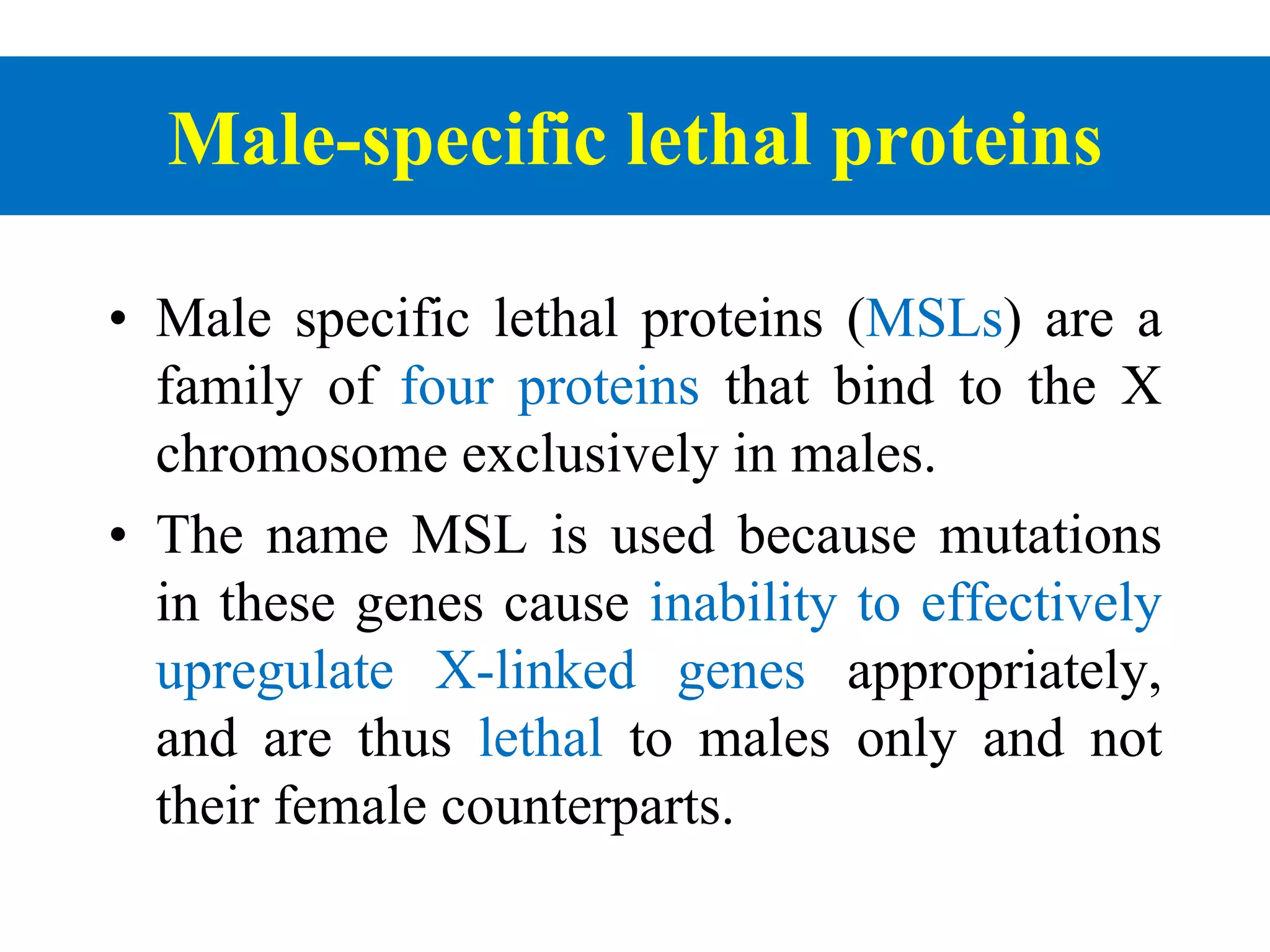
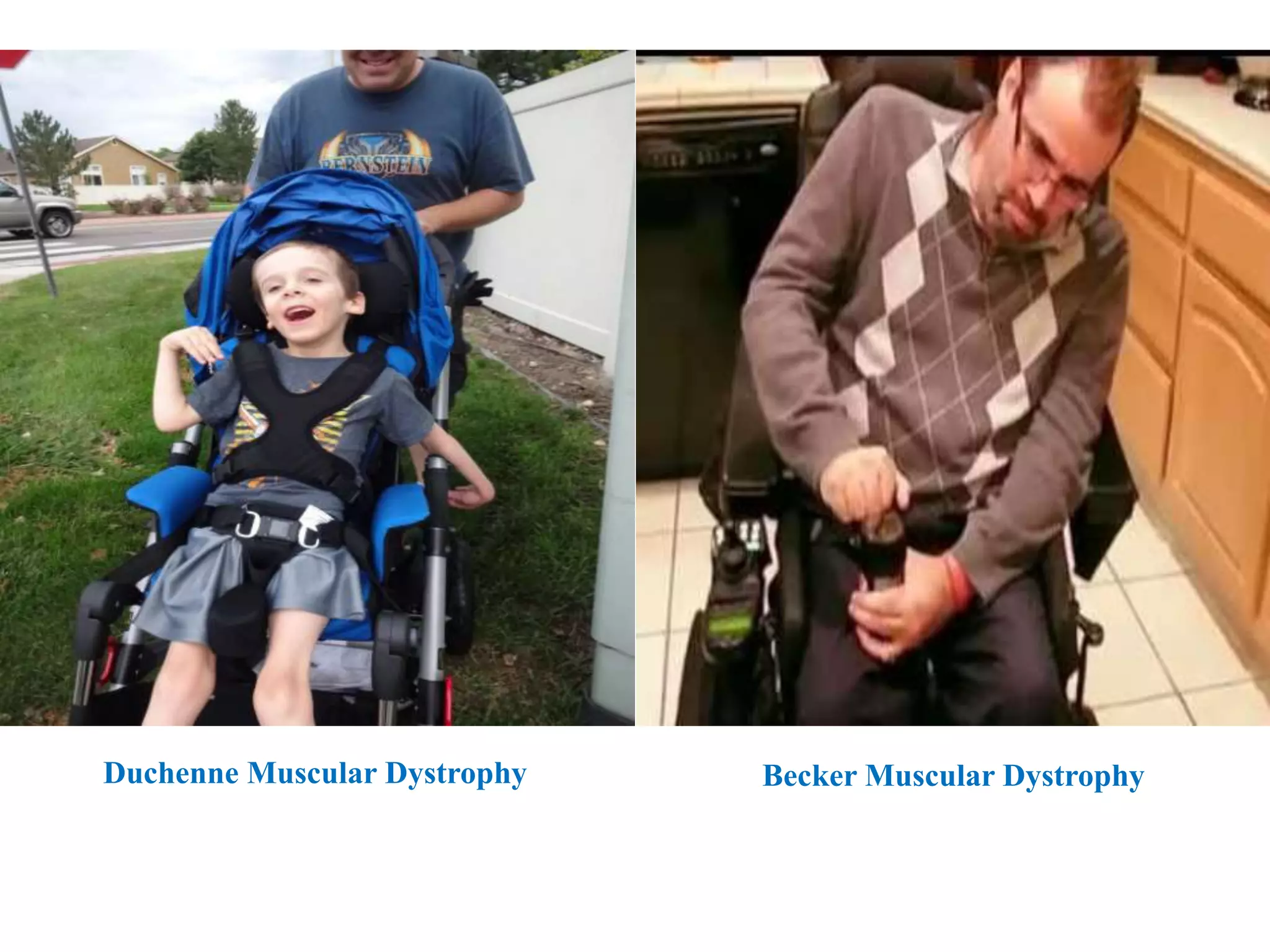
X inactivation is the process by which one of the two X chromosomes in female mammals is randomly inactivated. This occurs early in embryonic development through the expression of long non-coding Xist RNA from the future inactive X chromosome, which coats and silences that chromosome. Genes on the inactive X are transcriptionally silenced through epigenetic modifications, though some genes escape inactivation. The process ensures equal expression of X-linked genes between males and females through dosage compensation.