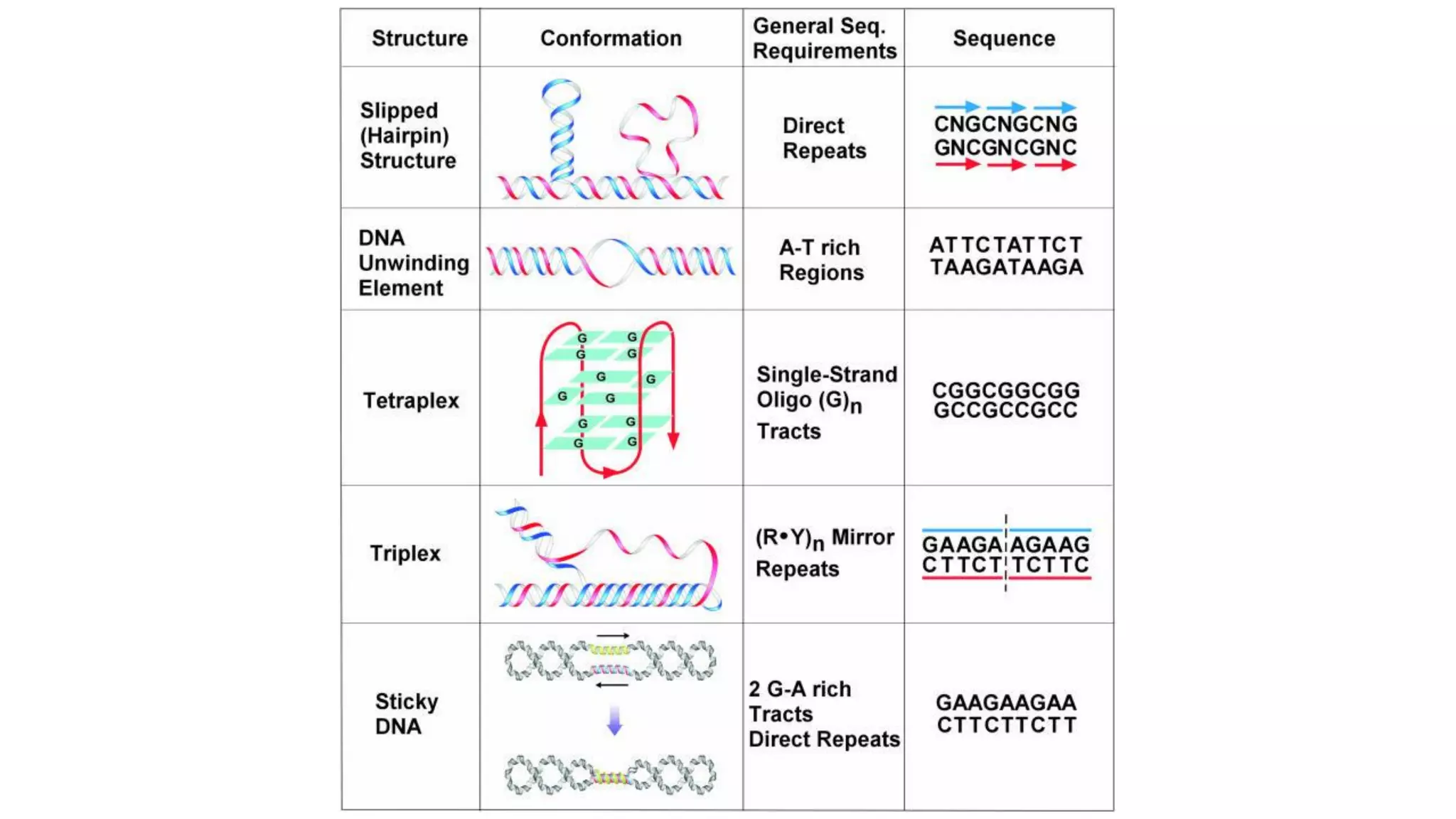
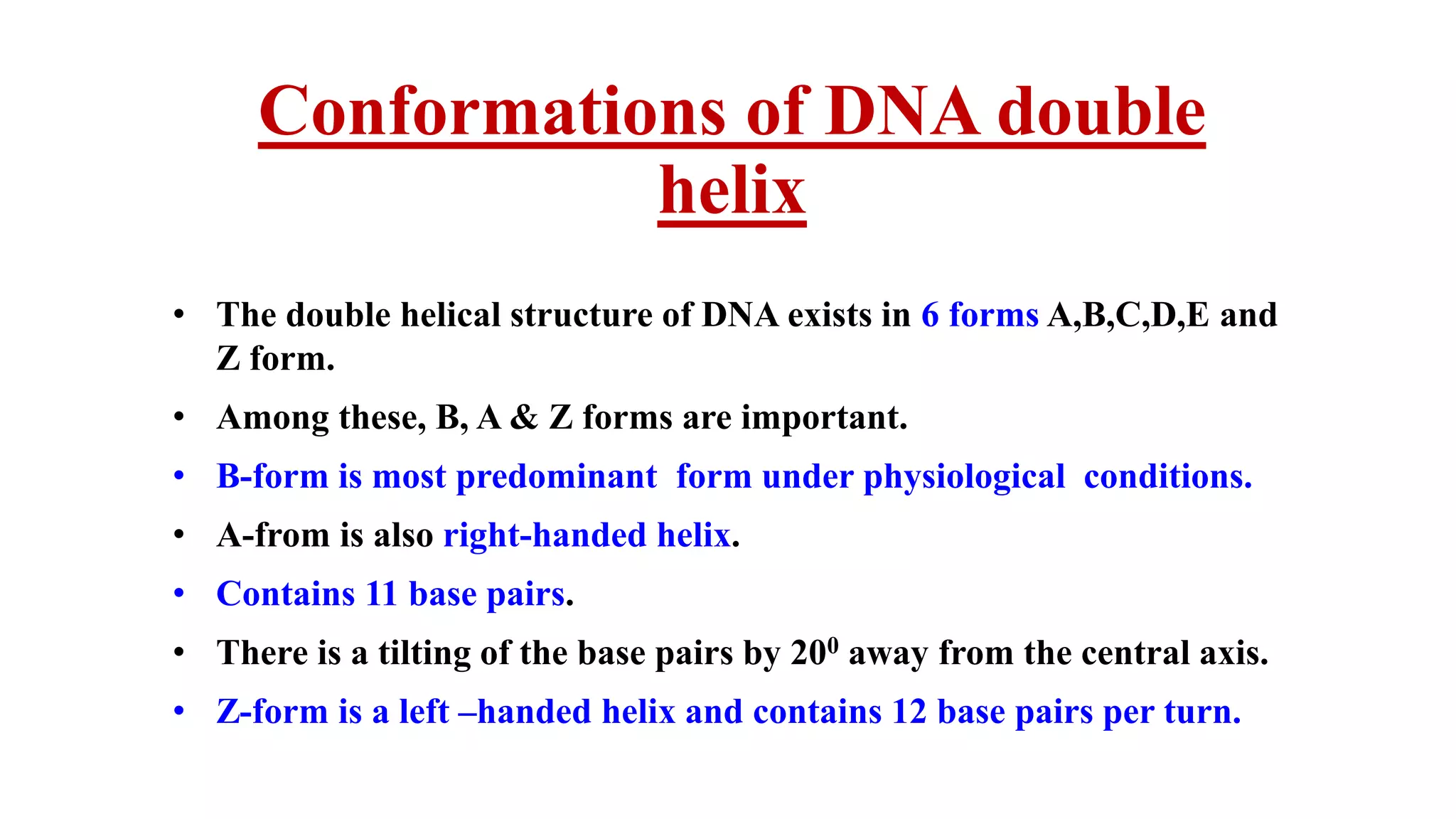
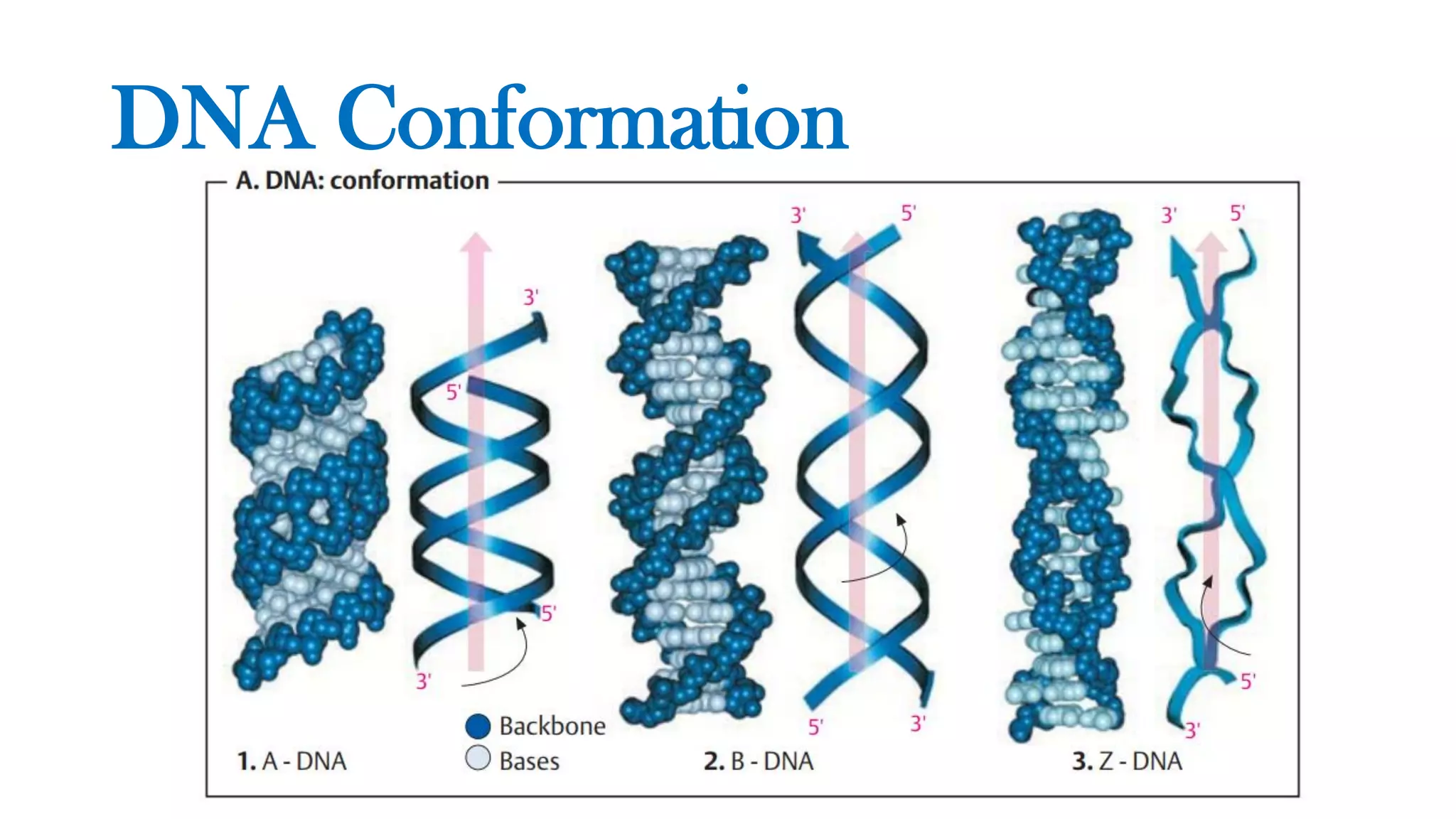
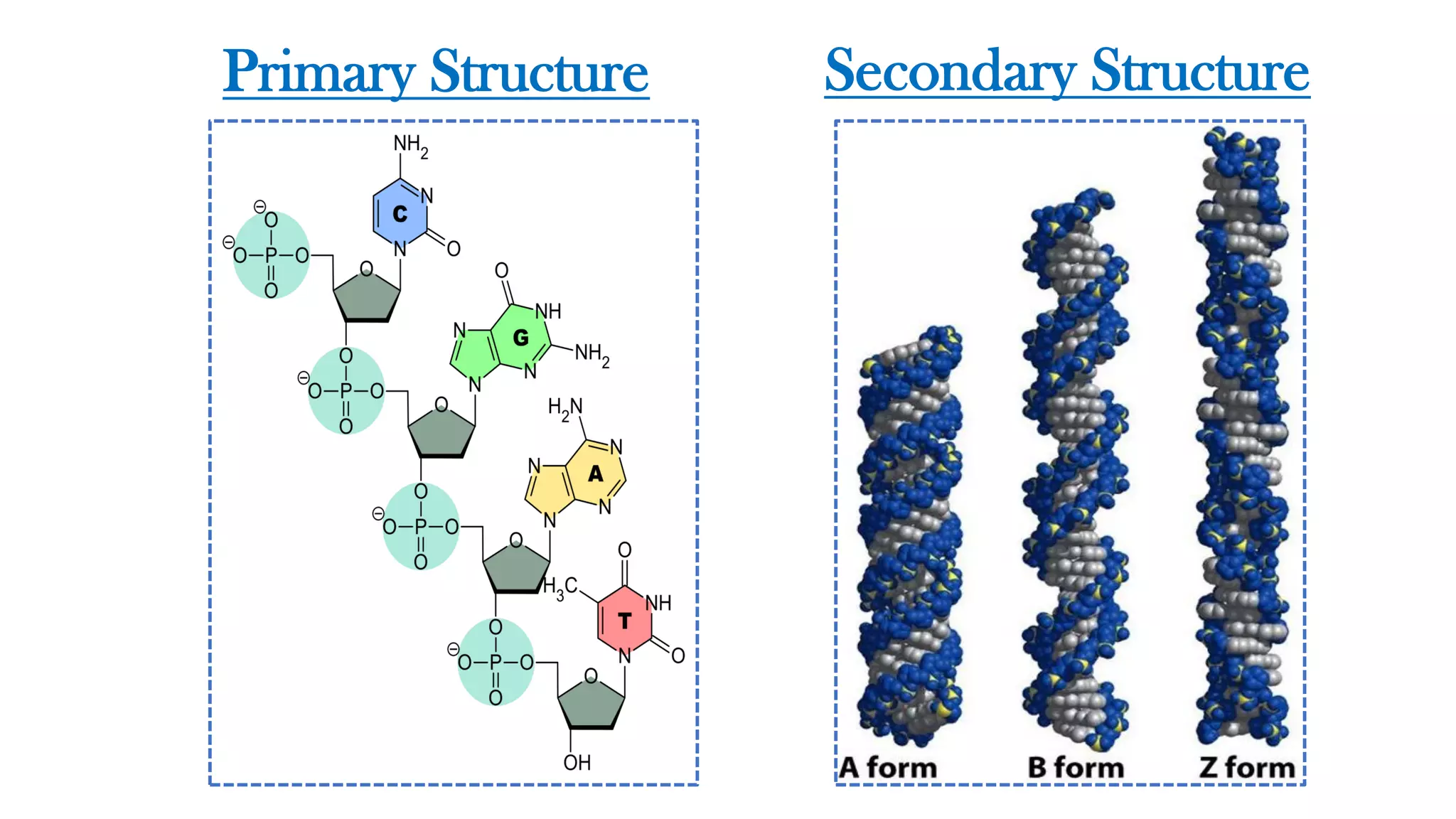
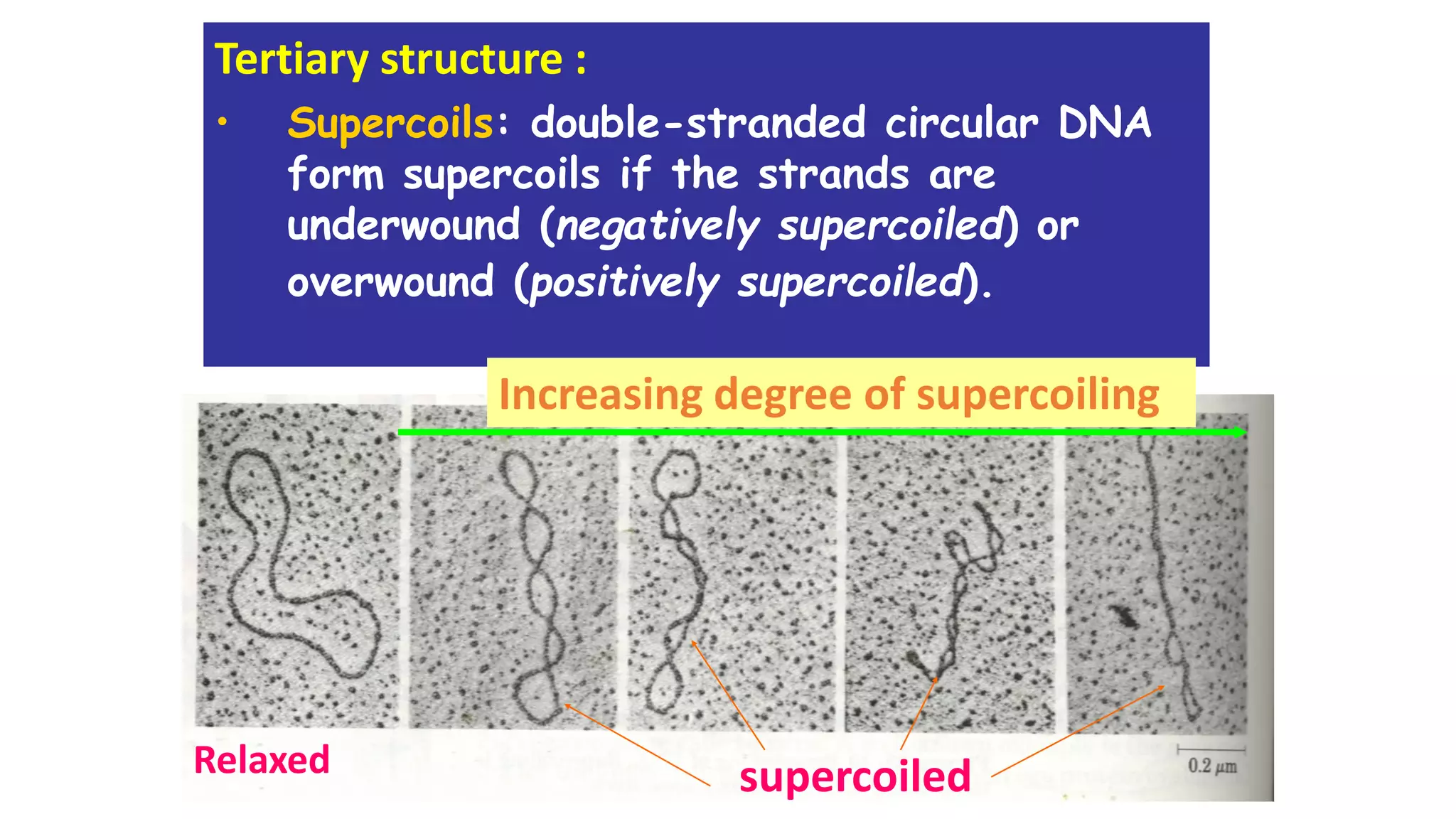
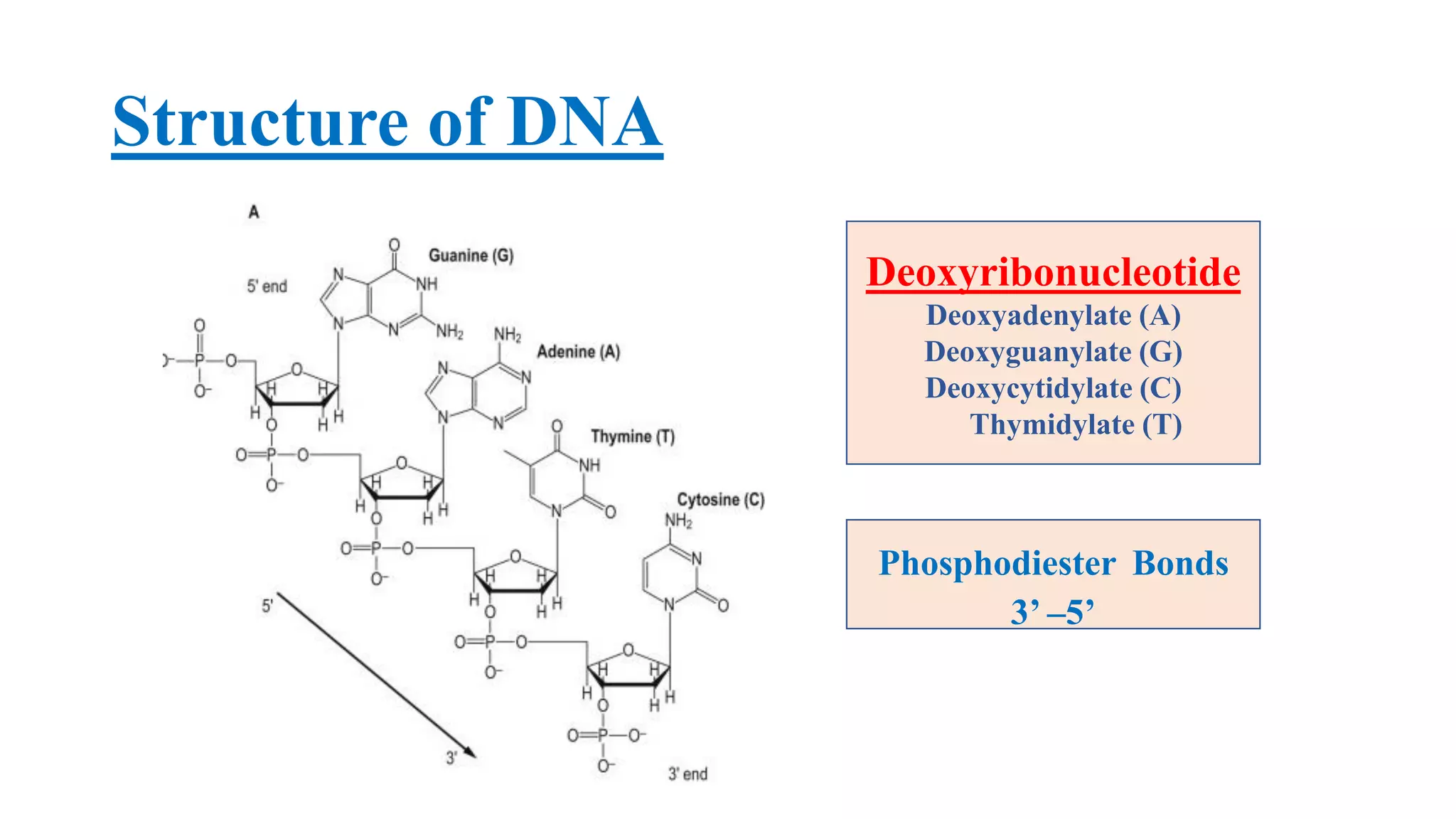
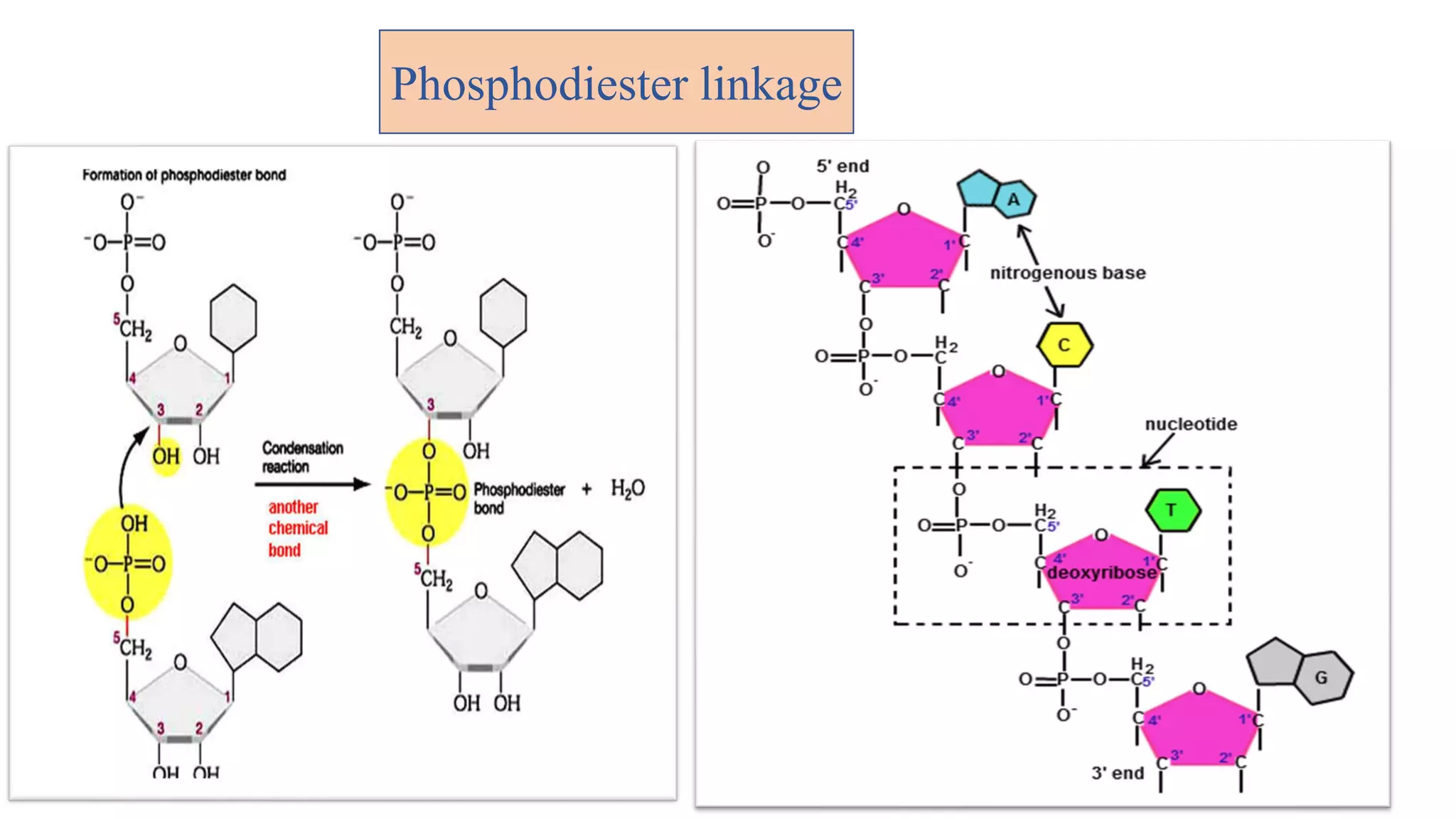
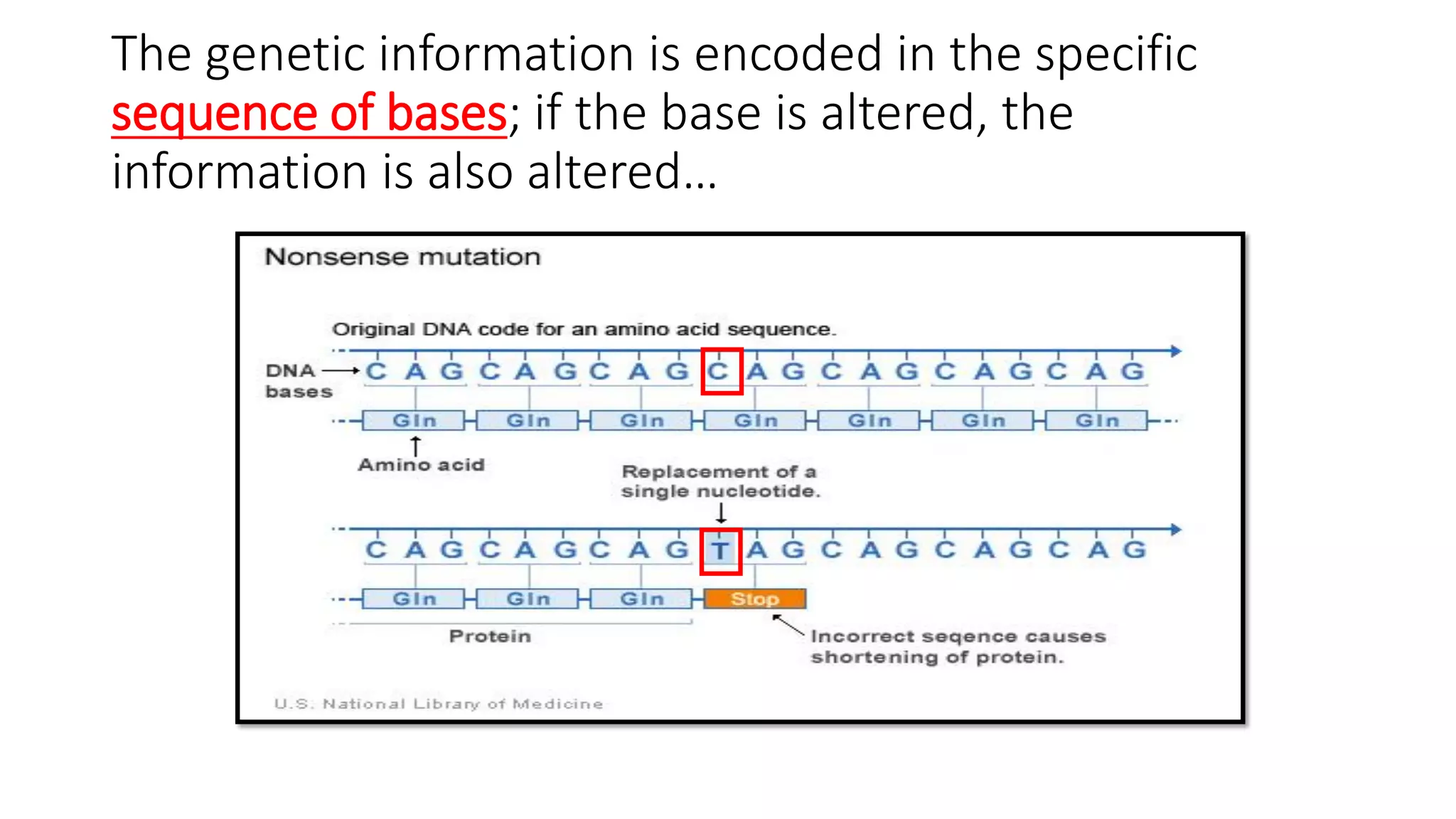
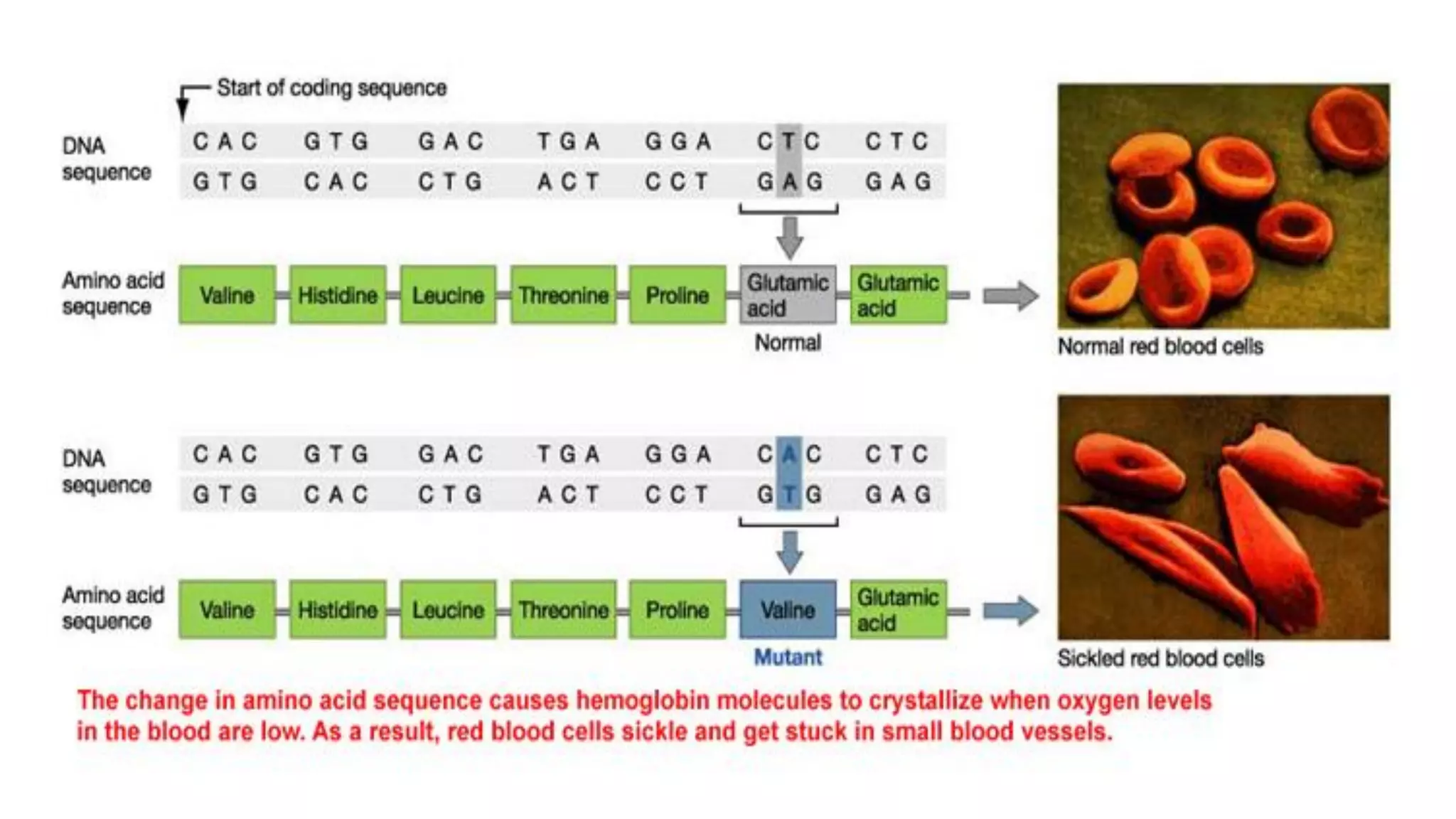
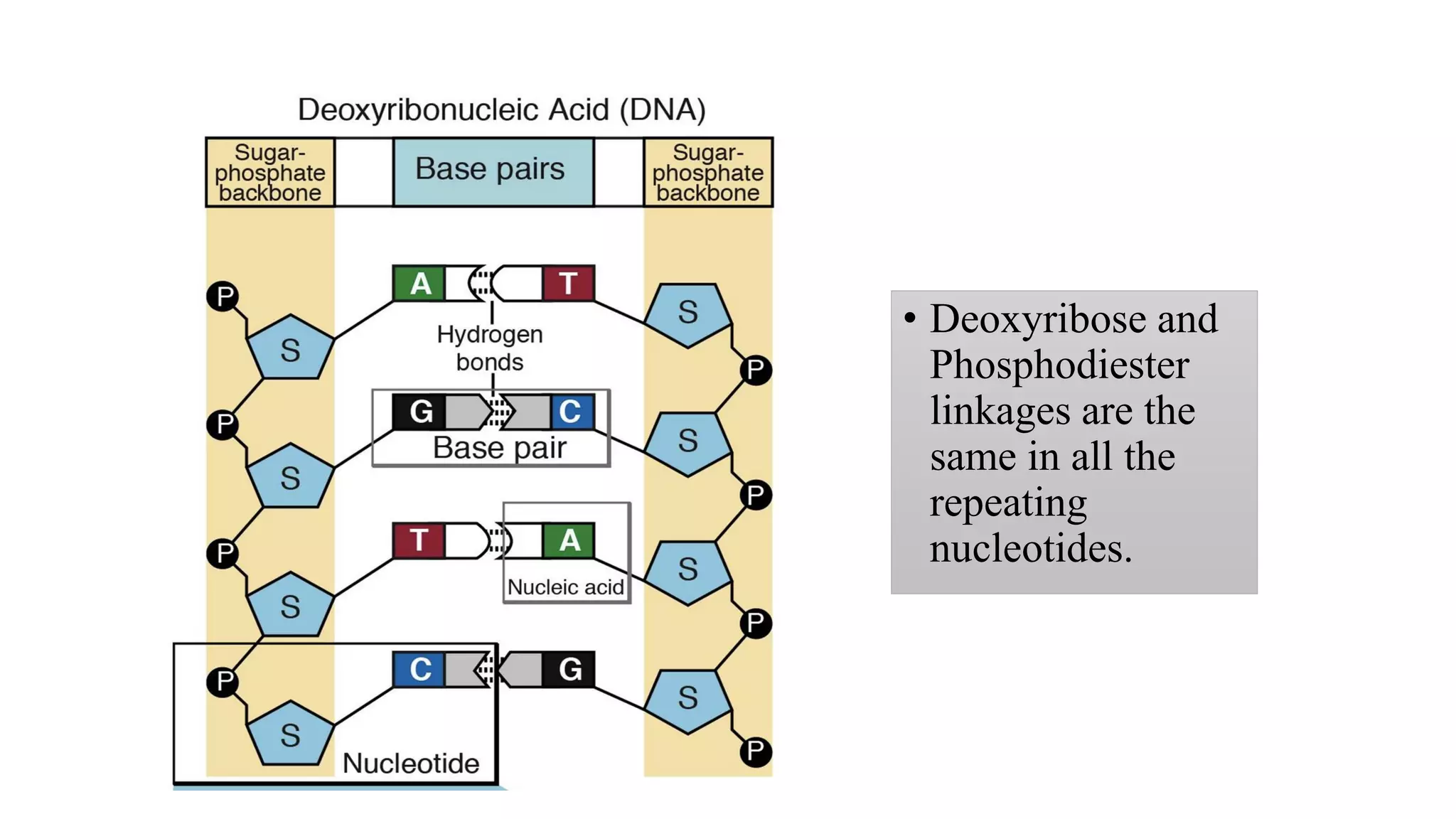
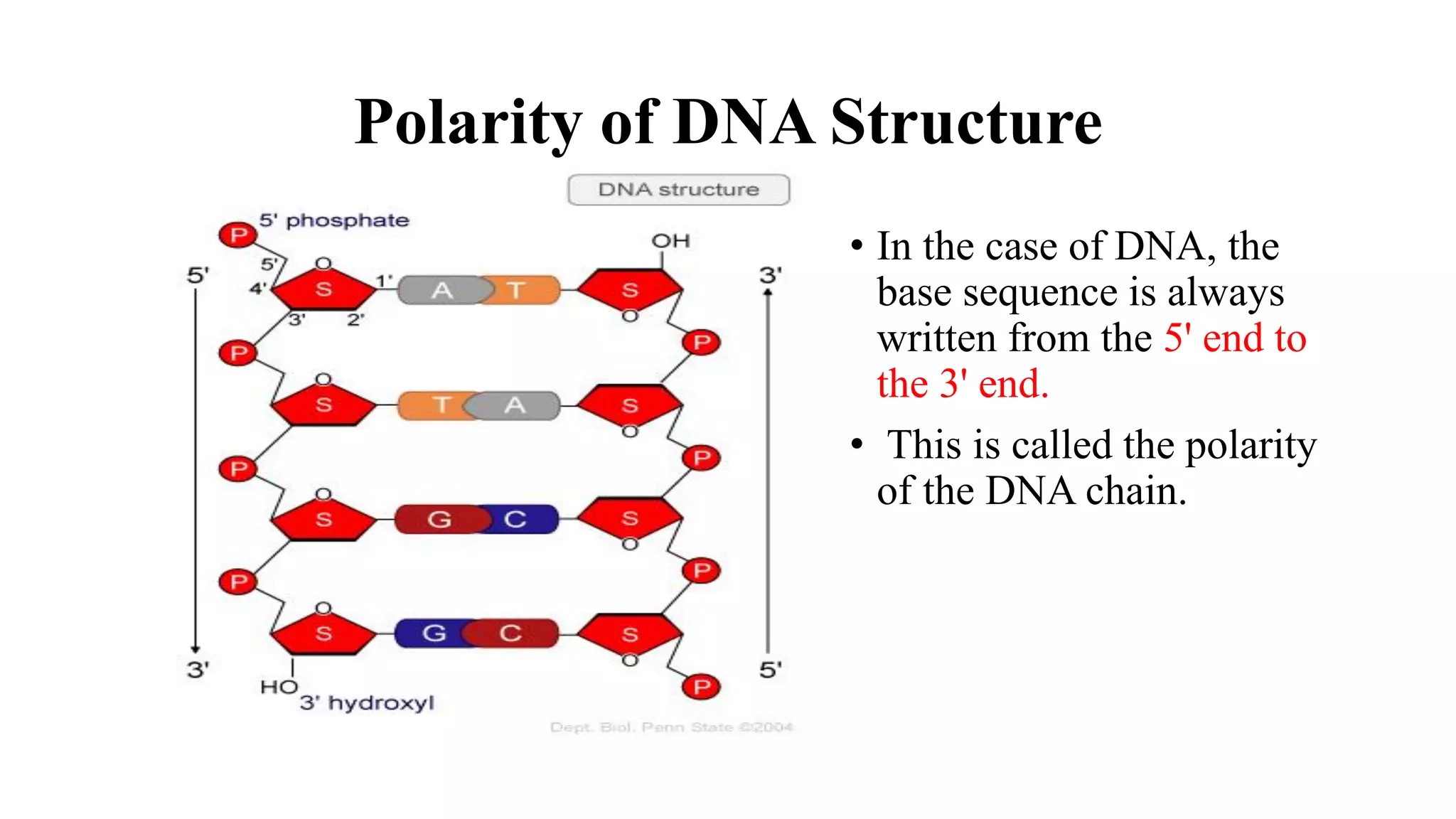
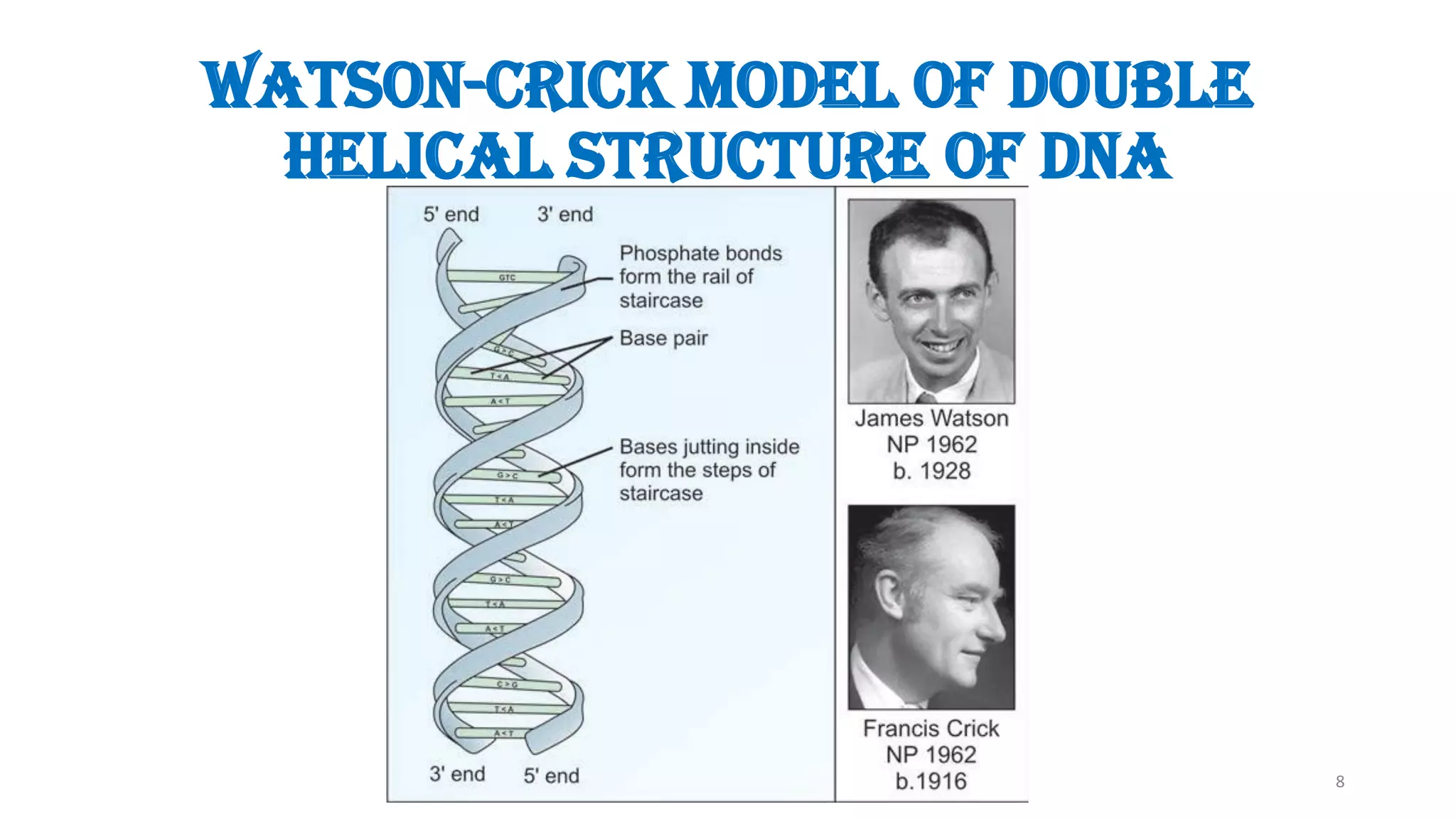
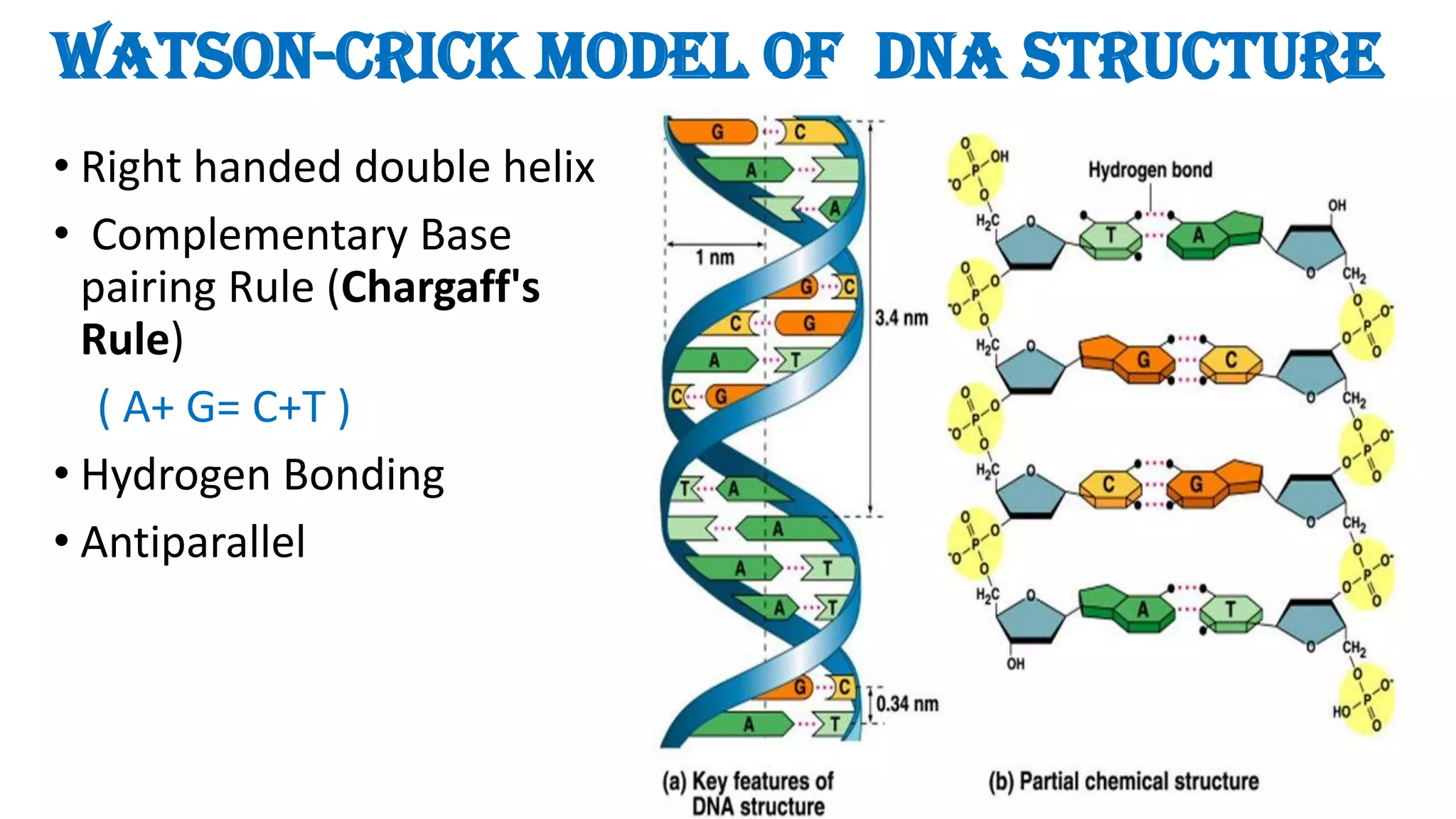
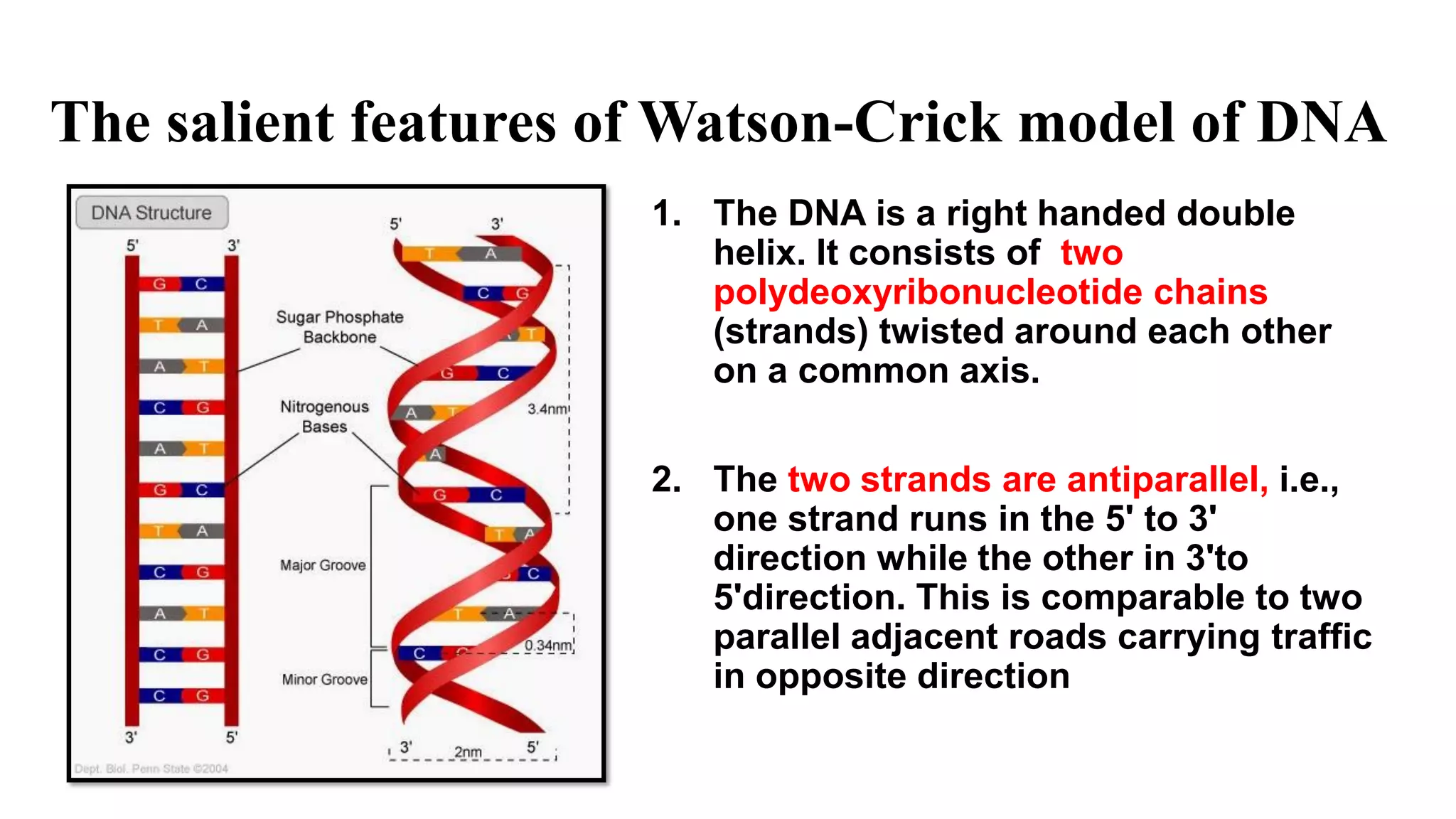
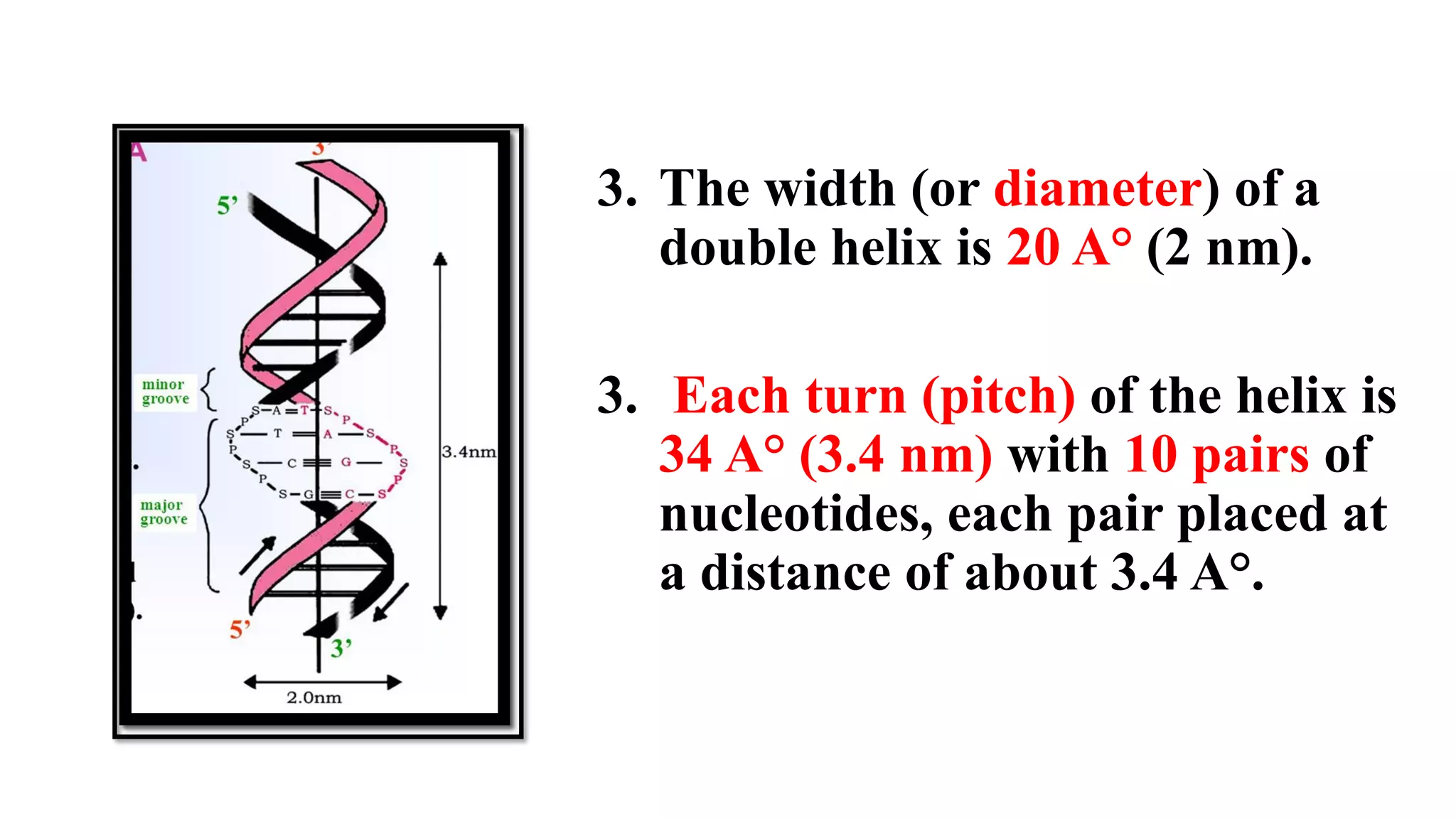
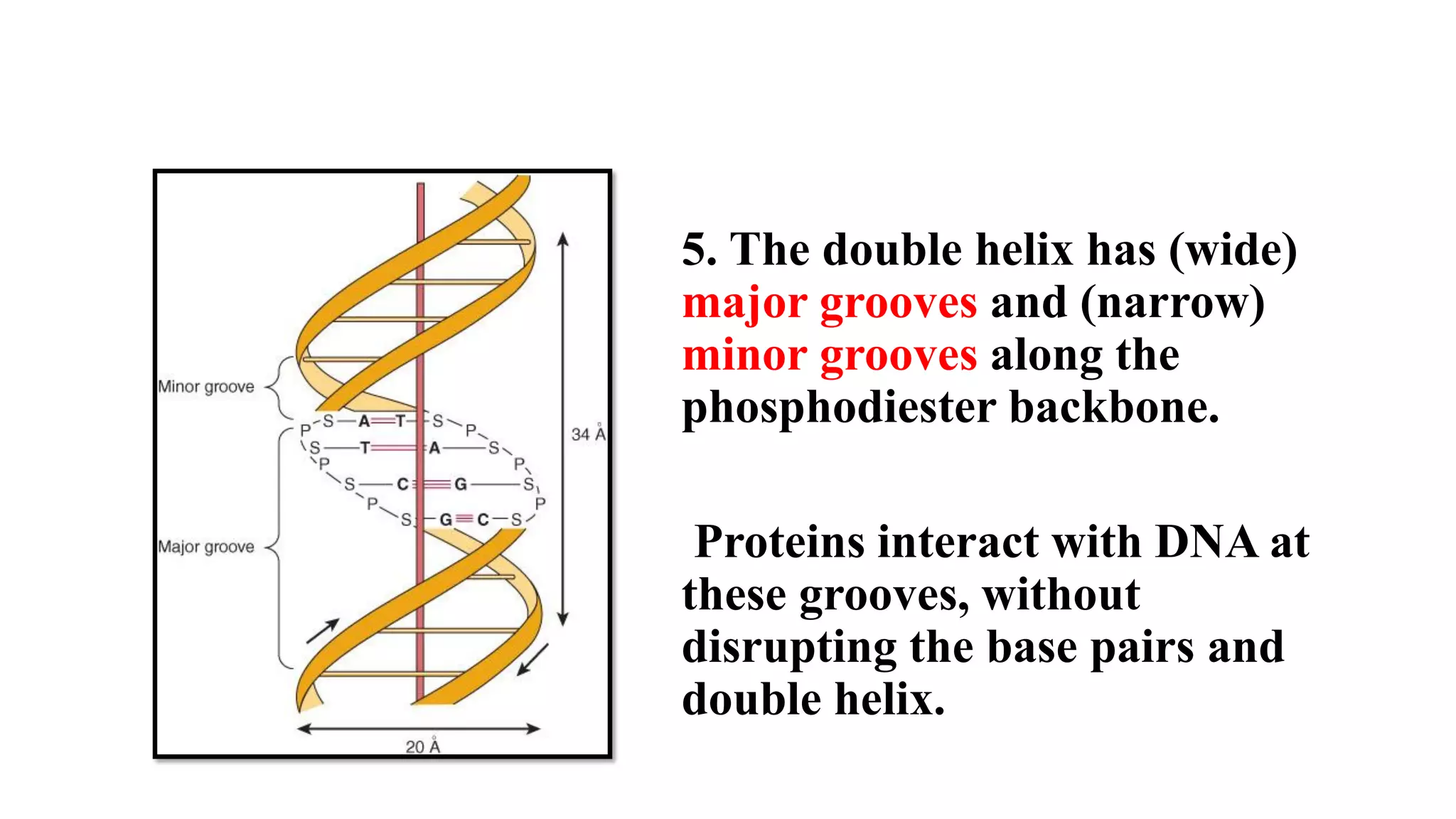
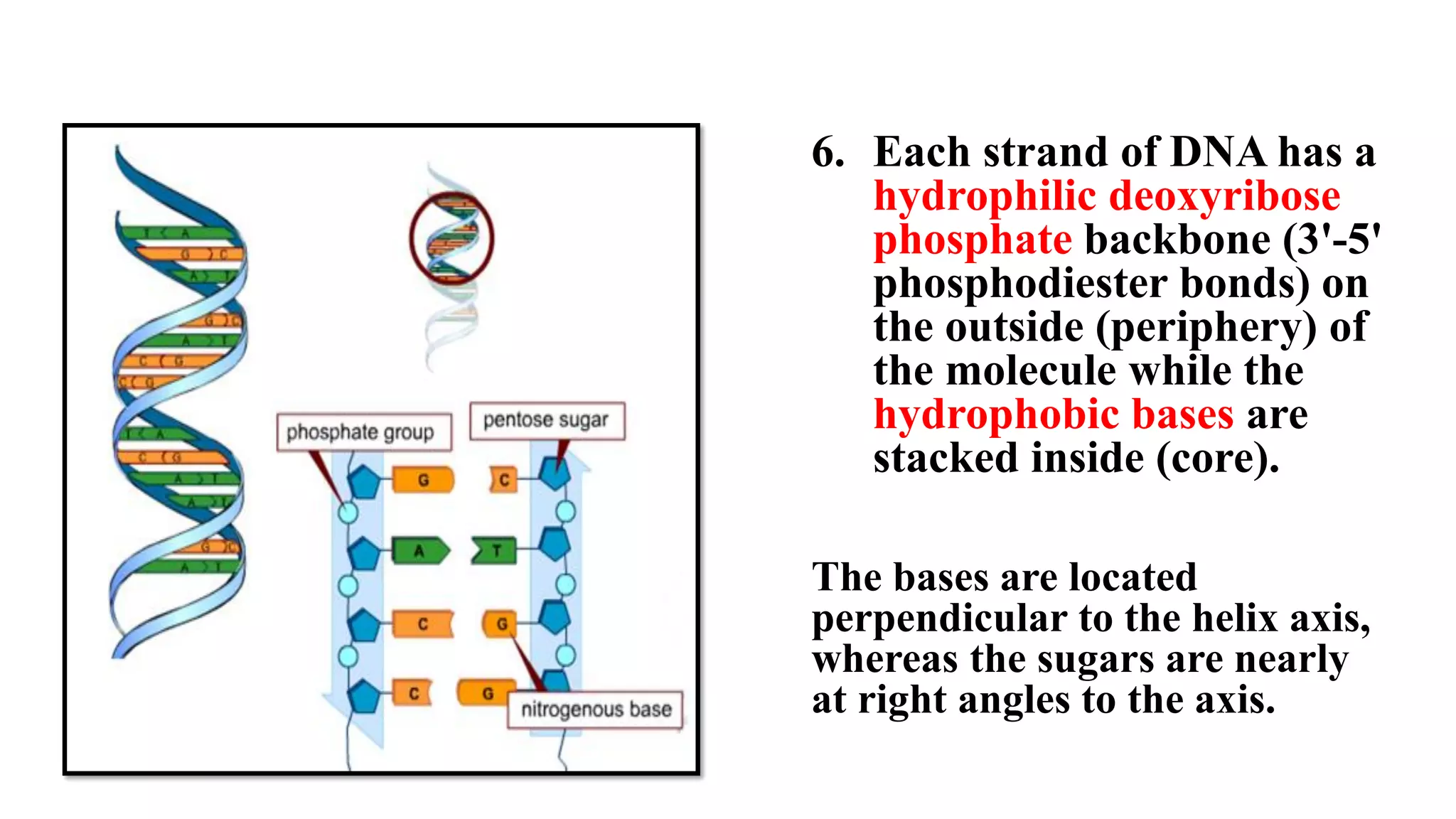
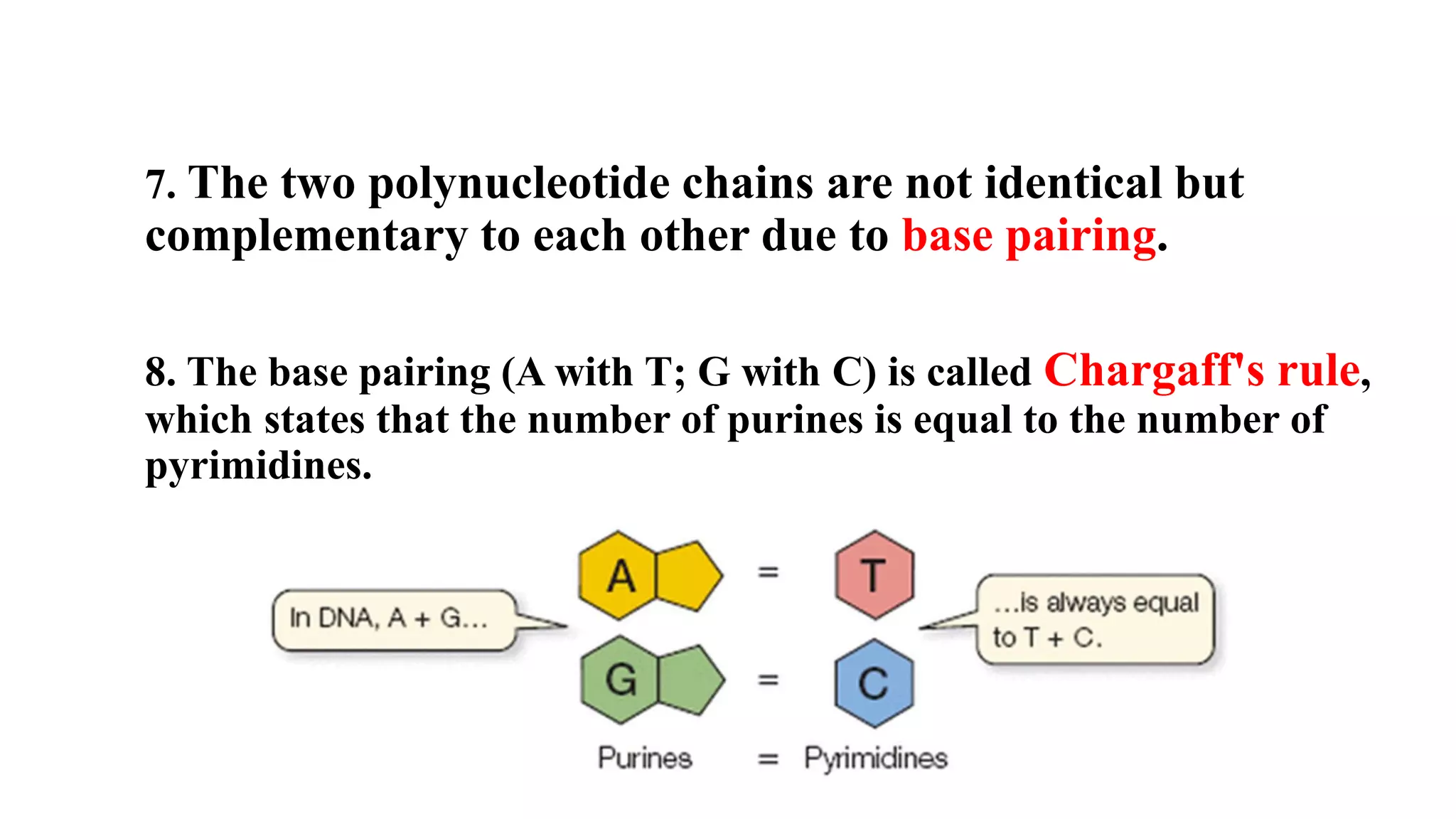
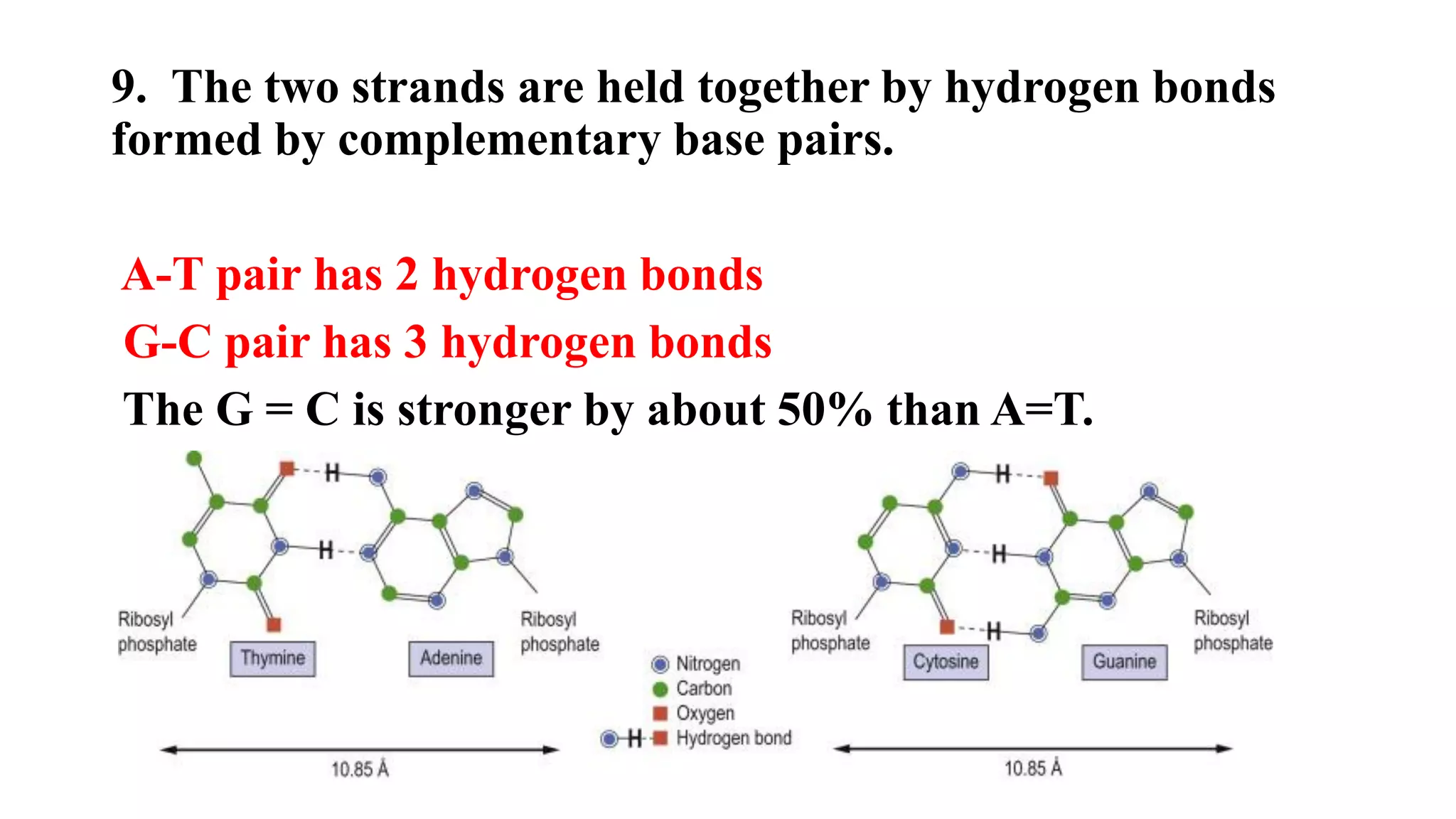
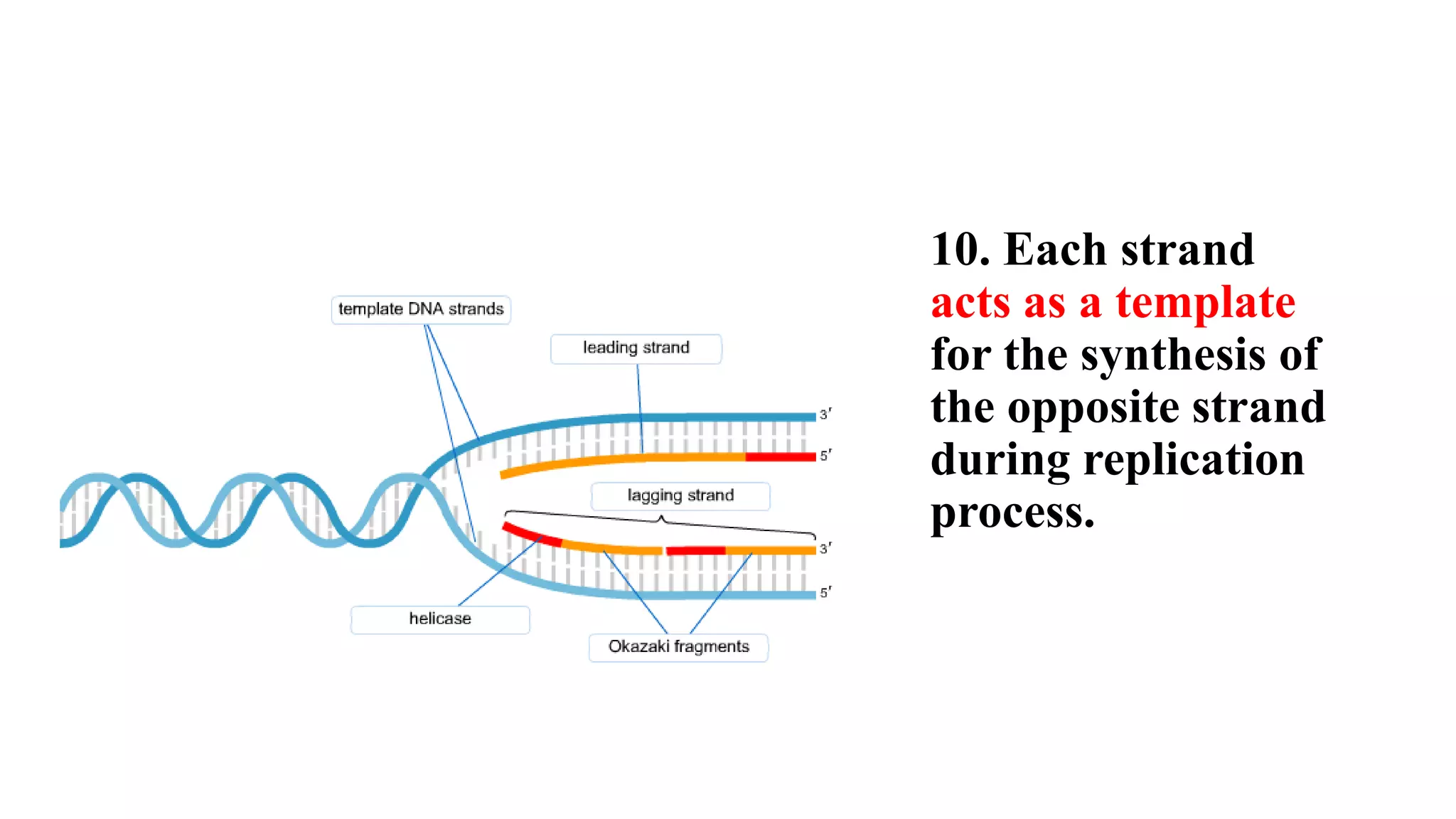
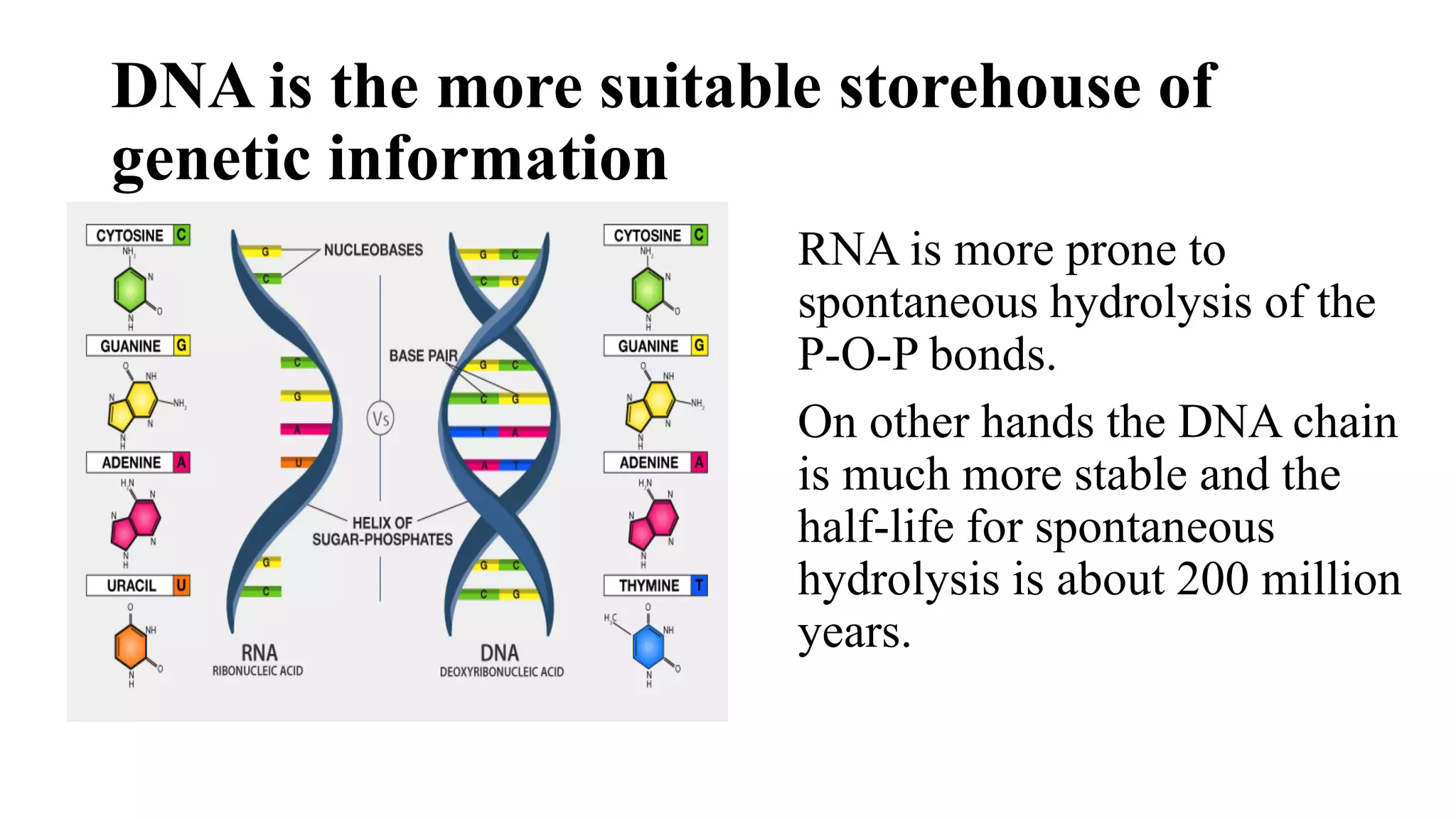
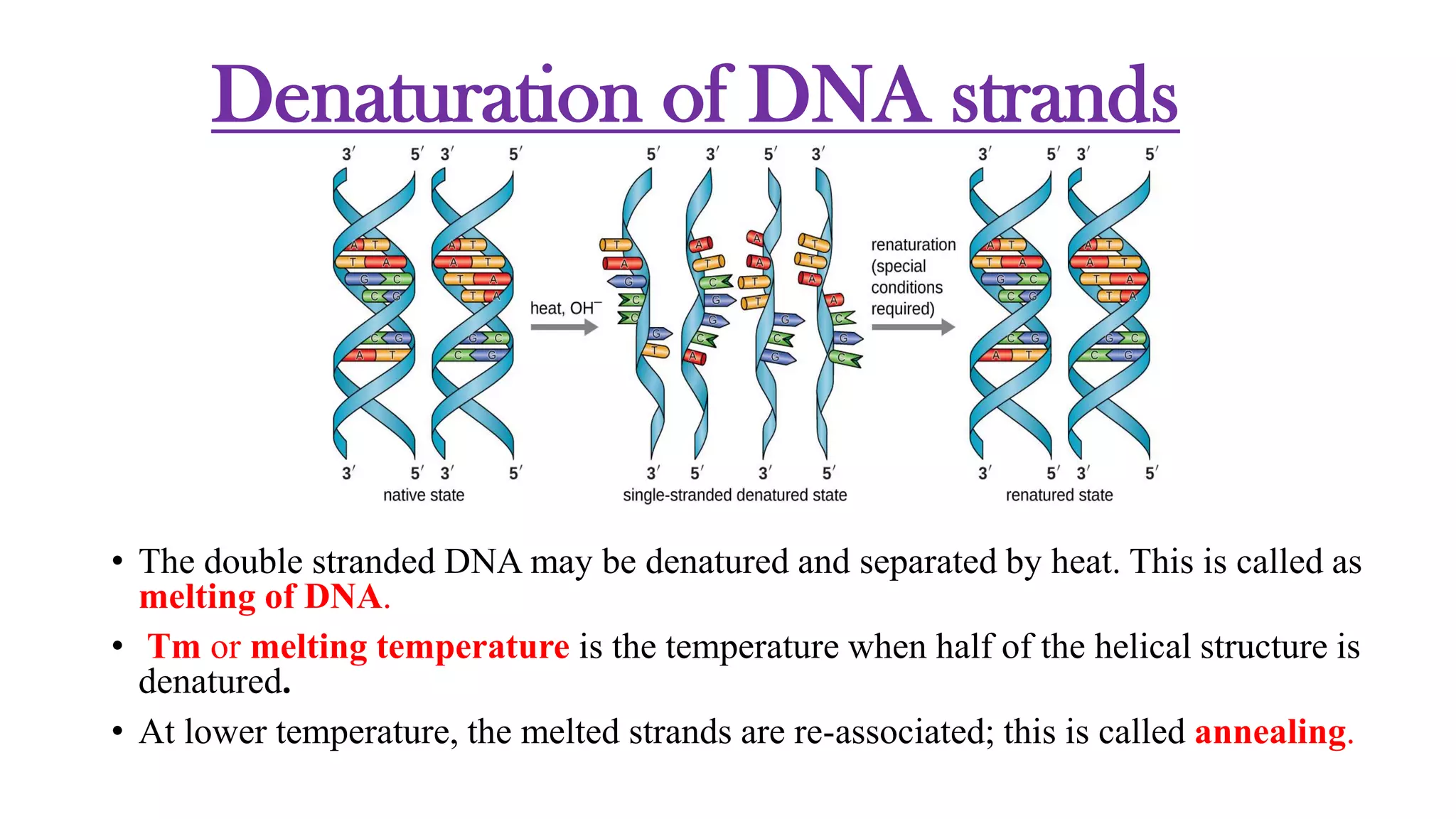
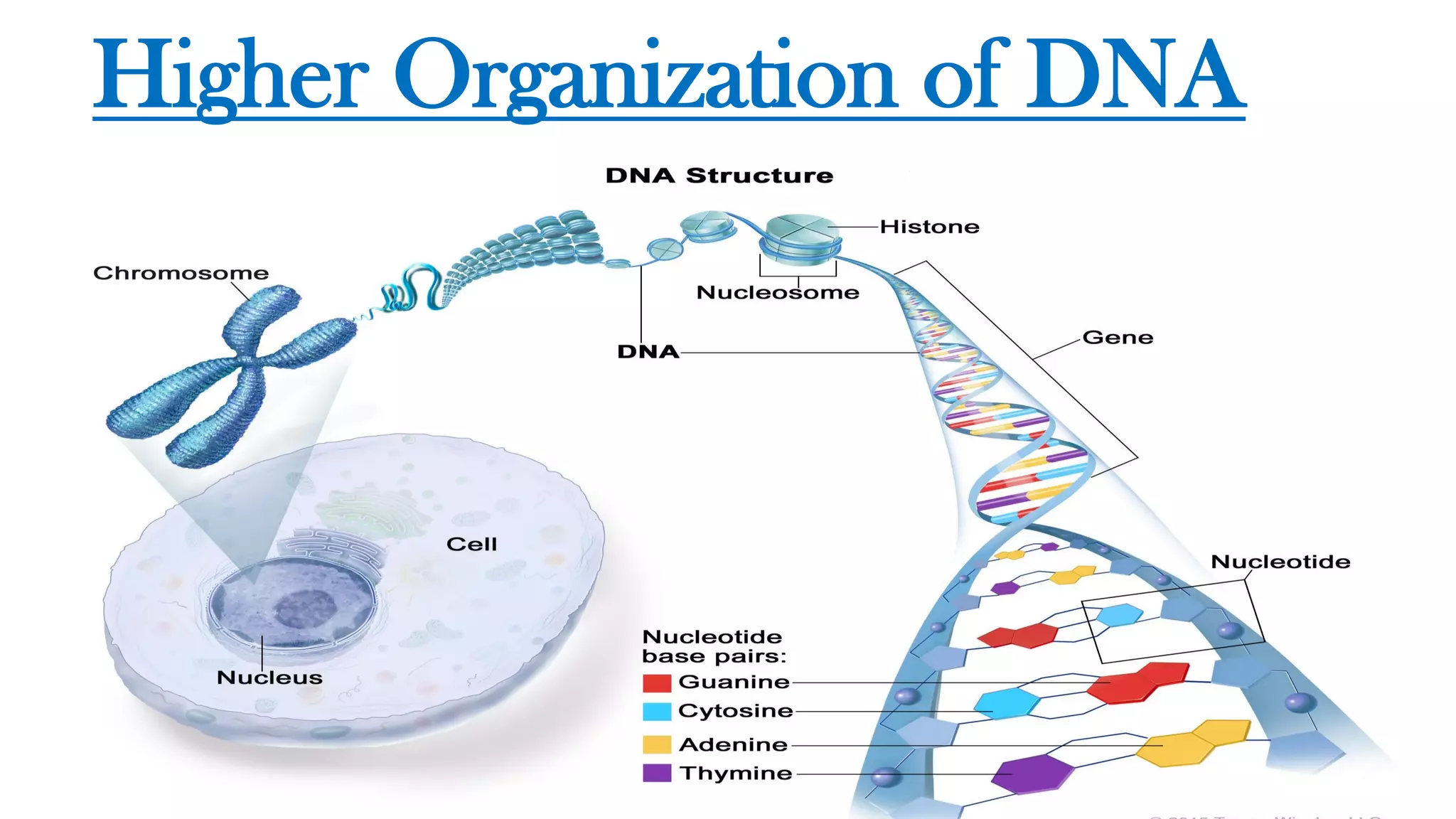
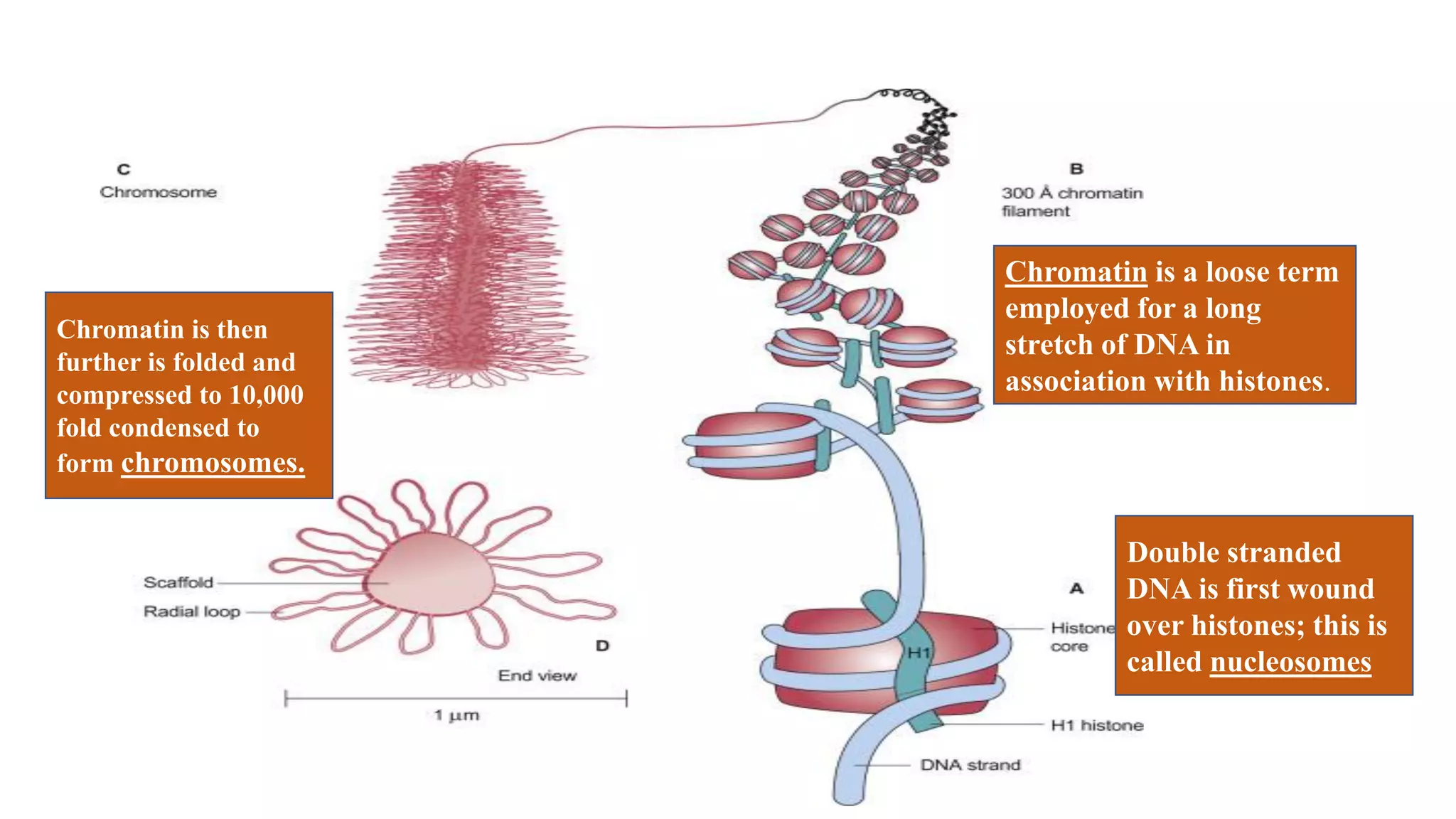
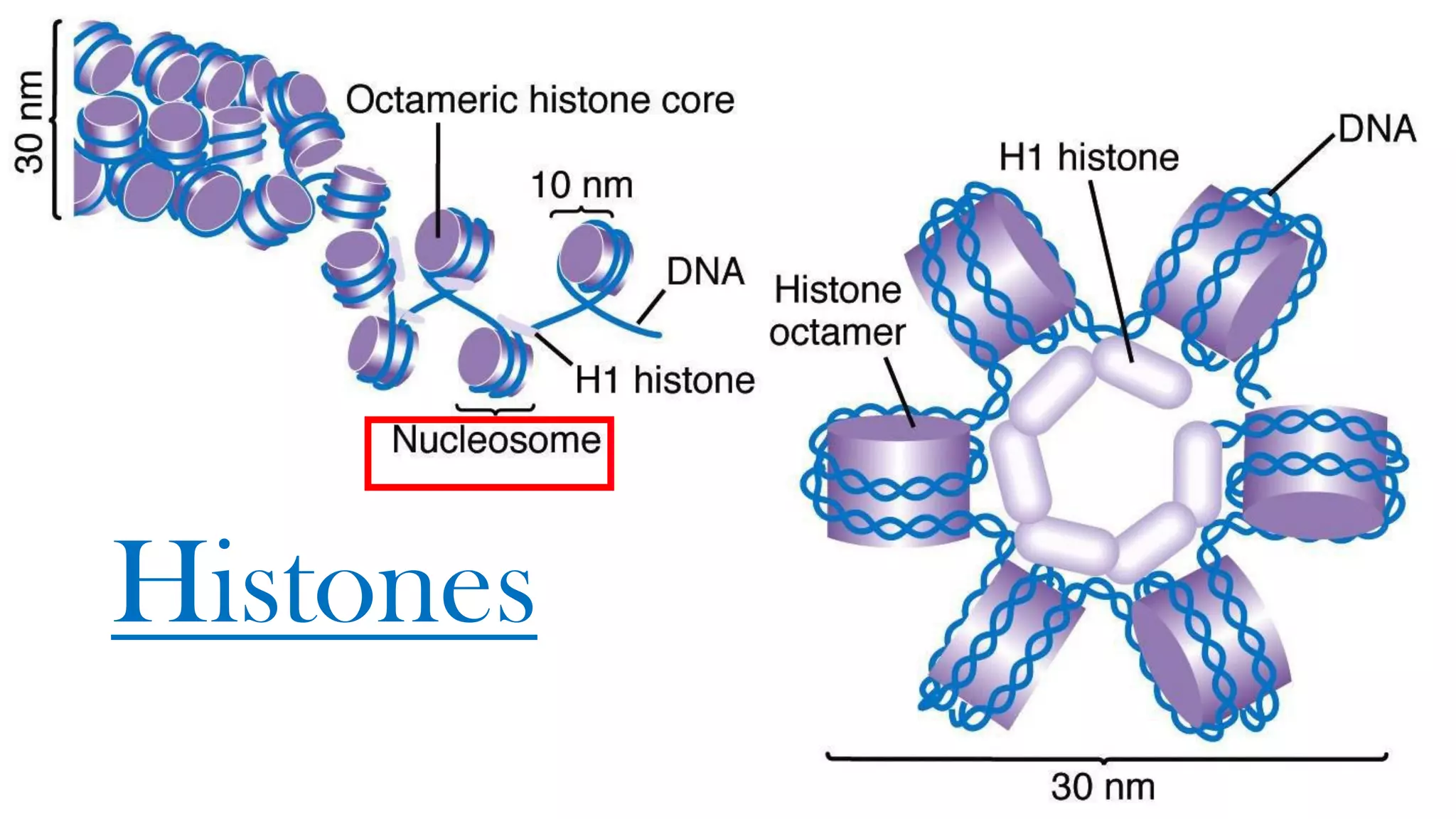
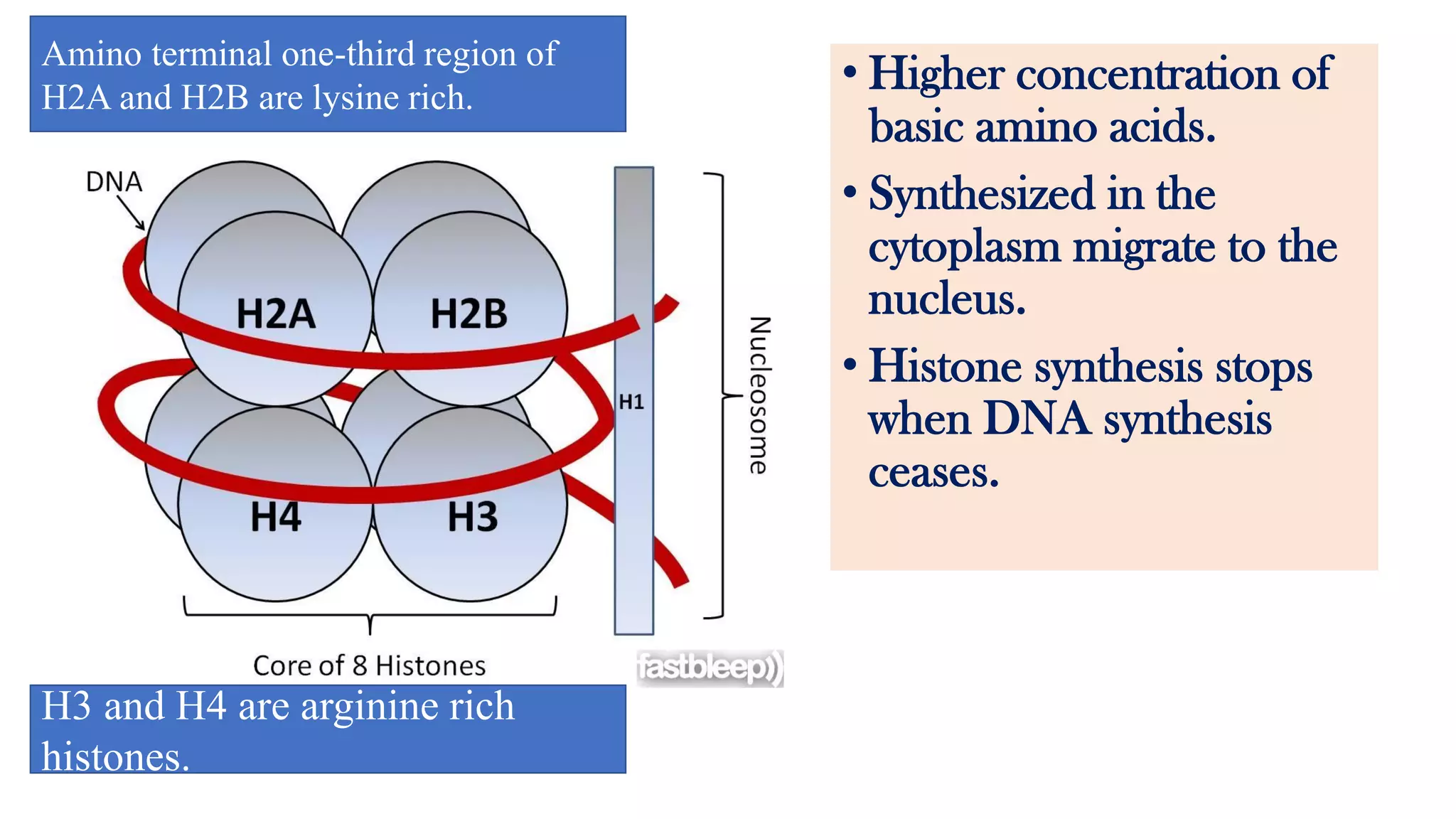
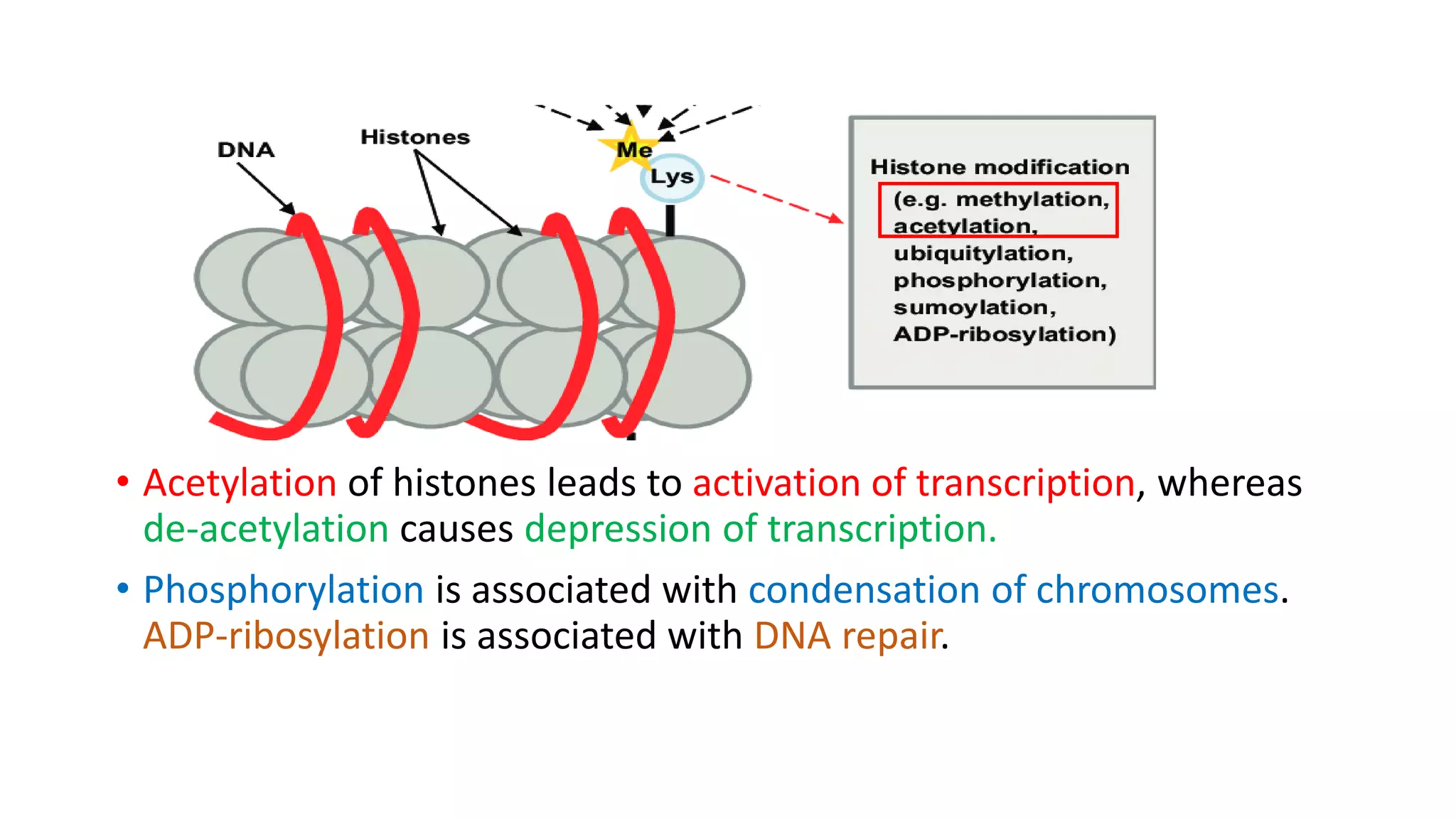
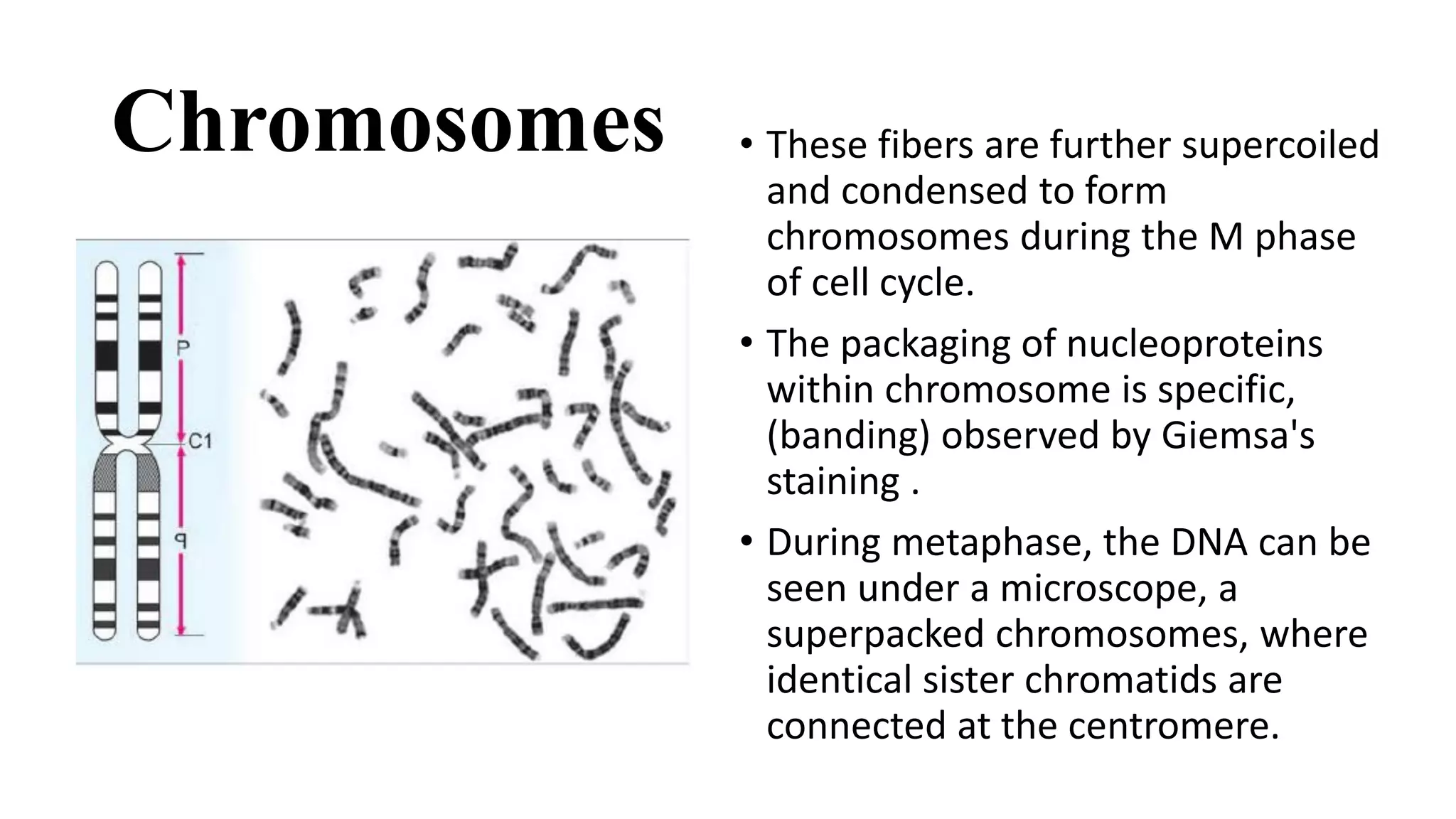
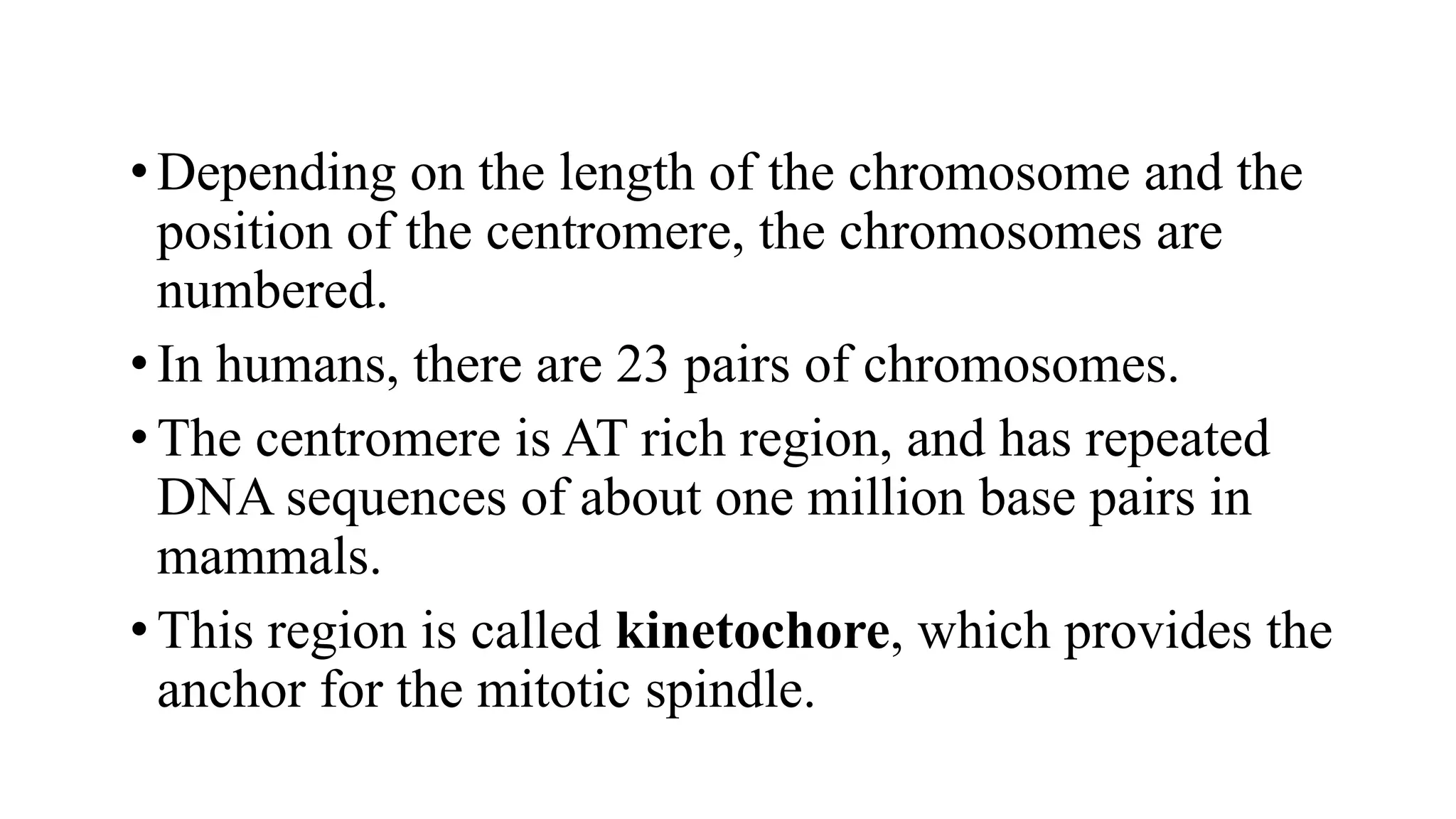
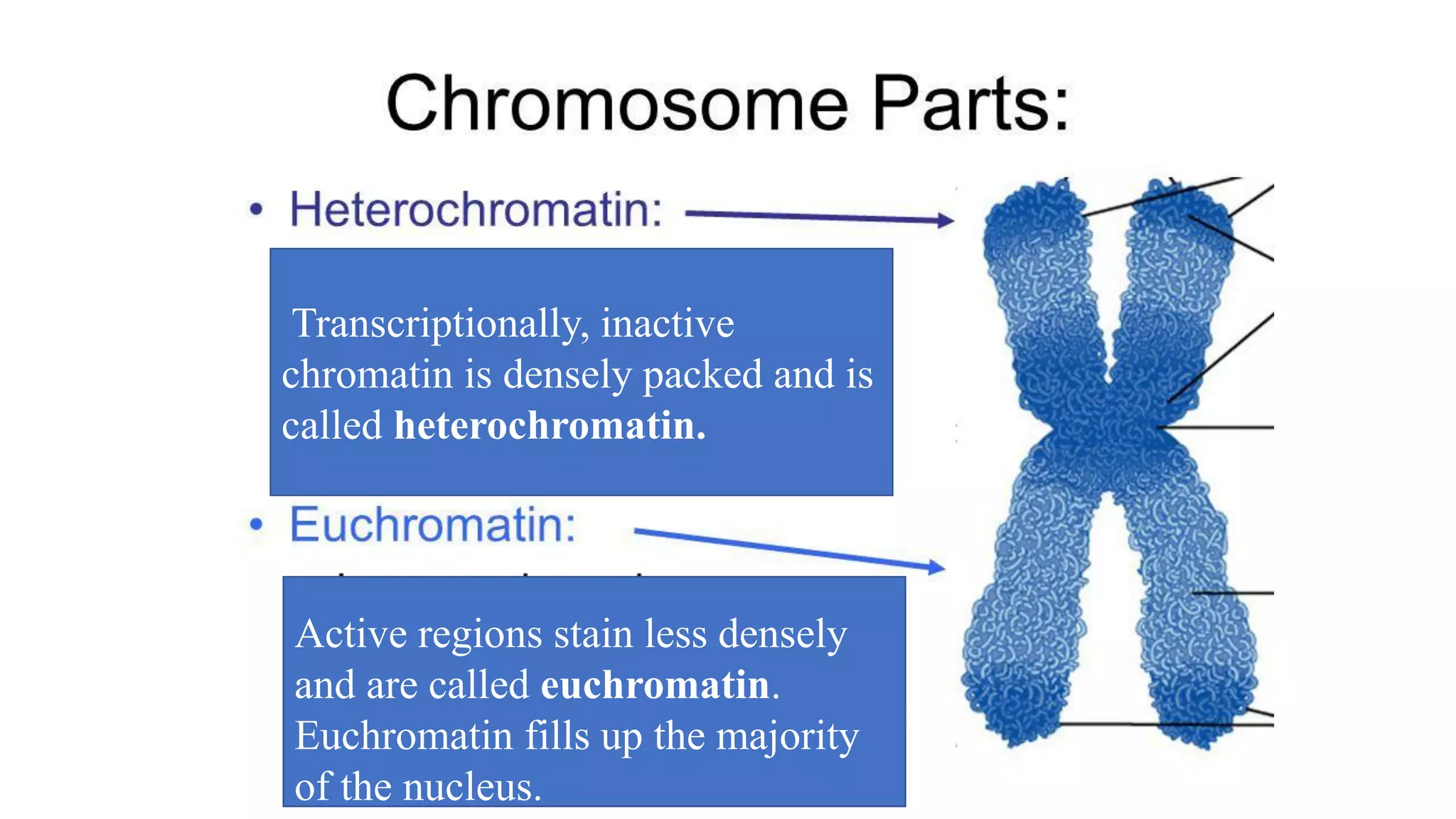
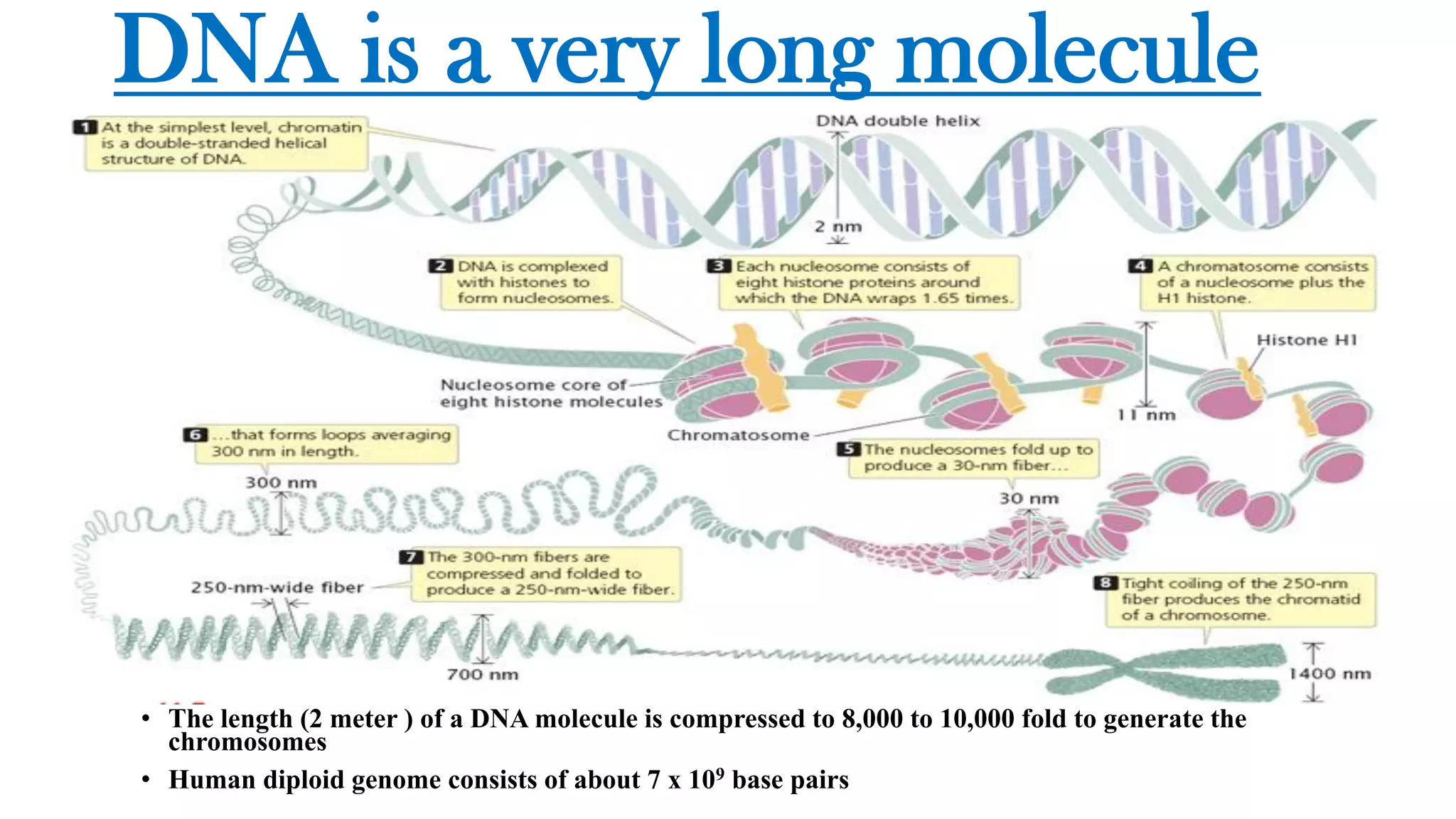
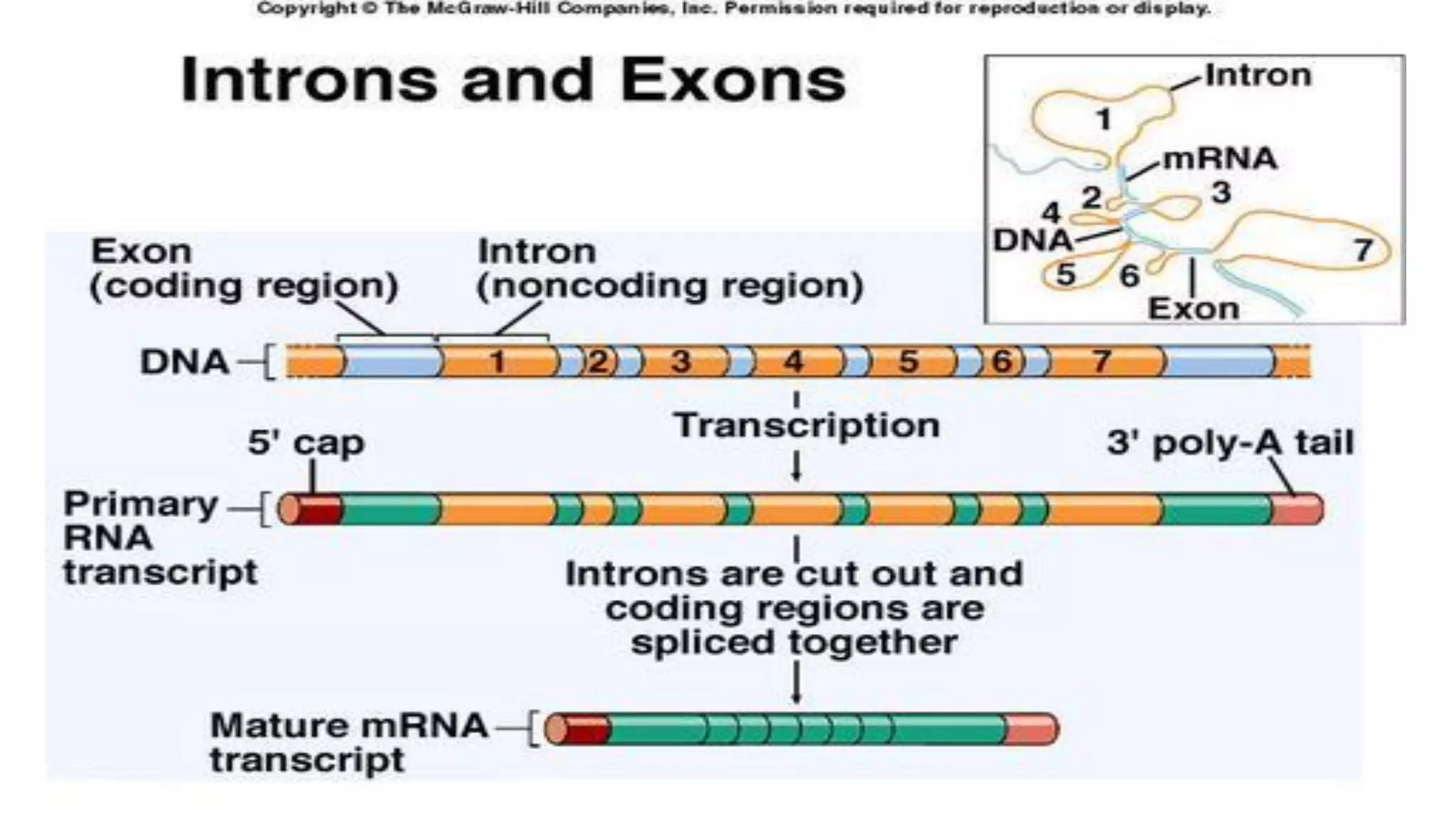
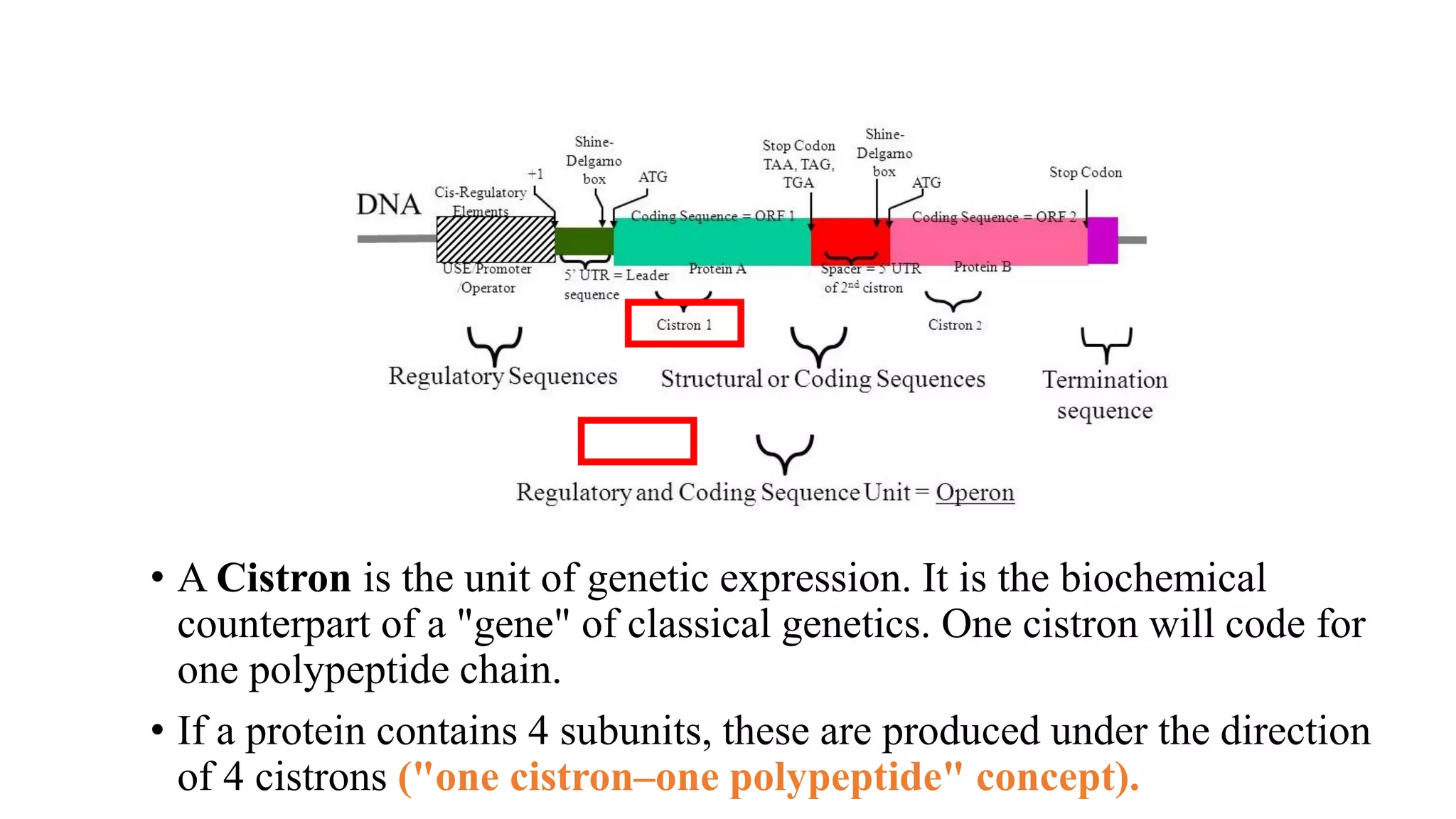
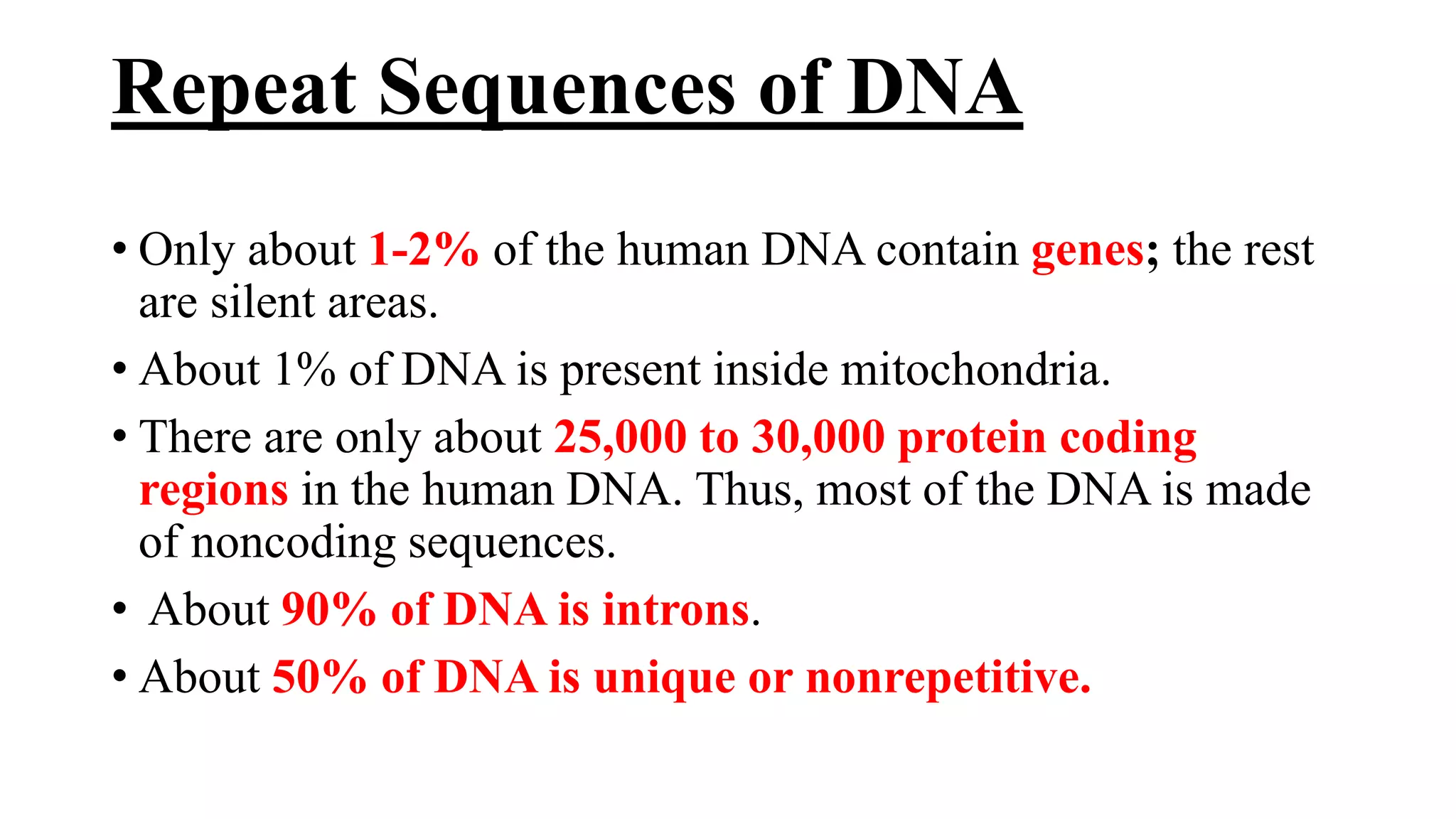
The document details the structure and function of DNA, including its double helical form as proposed by the Watson-Crick model, which highlights the antiparallel strands, base pairing rules, and the stability of the DNA molecule. It also explains the organization of DNA into chromatin and chromosomes, involving histones and various modifications that influence gene expression. Furthermore, it discusses the conformations of DNA, such as the different helical forms and the significance of repeat sequences within the genome.

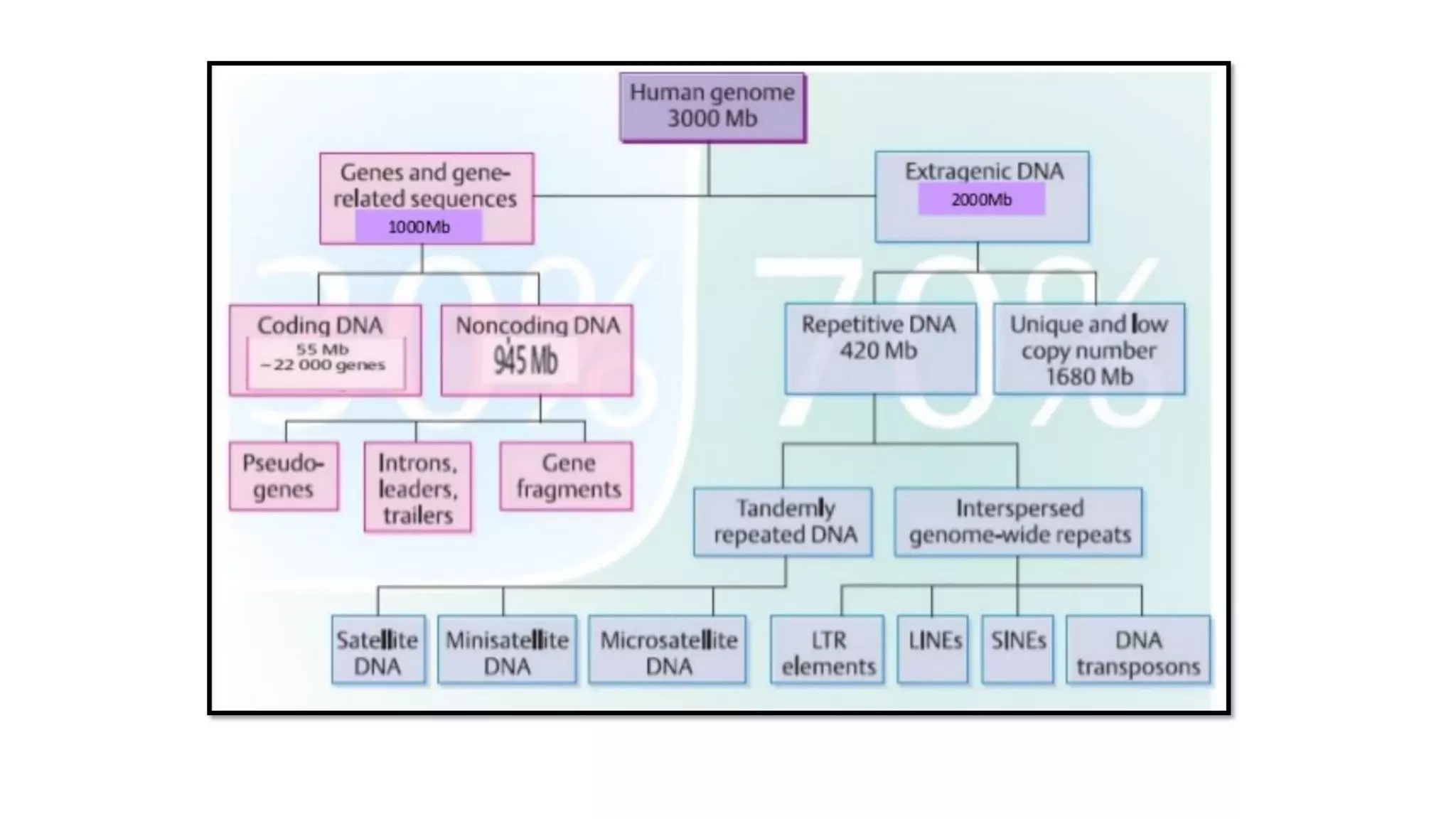
![• About 30% of the genome consists of repetitive
sequences; they contain 5- 500 base pairs repeated many
times.
[regions are clustered mainly in centromeres and
telomeres; but are seen randomly in other regions also.]
▪ Highly repetitive sequences
▪ Moderately repetitive sequences
▪ Long interspersed repeat sequences (LINEs)
▪ Short interspersed repeat sequences (SINEs).](https://image.slidesharecdn.com/dnachemistry-structurefuctionsanditsorgainization-200615110011/75/Dna-chemistry-structure-fuctions-and-its-orgainization-35-2048.jpg)
![• LINE are 6-7 kbp length and are repeated about 50,000 times.
• SINE are 100-300 bp in length, but are repeated 100,000 times.
[ One such sequence, the Alu family, is repeated about 500,000 times,
and accounts for about 5% of total human DNA.]
Characteristics:
✓ These interspersed repeat sequences are mobile elements, and they
can jump from one region to another region of the genome.
✓They appear to be retroposons, i.e. they can move from one location
to another (transposition) through an RNA intermediate by the action
of reverse transcriptase.](https://image.slidesharecdn.com/dnachemistry-structurefuctionsanditsorgainization-200615110011/75/Dna-chemistry-structure-fuctions-and-its-orgainization-36-2048.jpg)