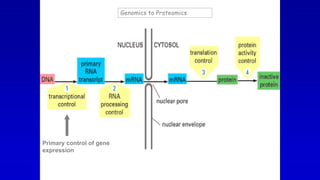
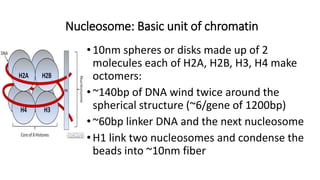
The document summarizes the organization of the human genome and genes. It discusses the general organization of the human genome including nuclear and mitochondrial genomes. It describes gene distribution and density in the nuclear genome. It provides details on the organization of different types of genes such as rRNA, mRNA, small nuclear RNA genes, overlapping genes, and multi-gene families. It also discusses various repetitive elements in the genome including SINEs, LINEs, microsatellites, and minisatellites. Finally, it covers topics like chromatin structure, histones, heterochromatin, euchromatin, and X-inactivation.




















![X-inactivation:
•Xq13: XIC X-inactivation centre; encodes large non-
coding RNA called XIST(X-inactivation-specific
transcript), only in case of inactive copy; XIST primary
transcript splicing & polyadenylation 17Kb
mature RNA; recruits certain proteins that organize
the chromatin into closed, transcriptionally inactive
conformation [essential for inactivation but not for
maintenance of inactivation]
•What are the molecular changes in X-Inactivation or
‘heterochromatization’ that is initiated by XIST that
propagates along the whole length?](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-22-320.jpg)

![X-inactivation as mechanism of gene dosage
compensation: Total of partial?
• One of the X is inactivated over most of the regions
except;
• Two Pseudo-autosomal regions; PAR1 on Xp/Yp,
and PAR2 on Xq/Yq have functional homologues,
all PAR1 and some of PAR2 genes are found to
escape inactivation
The pseudoautosomal regions at the tips of Xp and Yp are identical, as are
those at the tips of Xq and Yq. The non-recombining male-specific region on
the Y chromosome (MSY) and the equivalent, X-specific, region on the X
chromosome are rather different in sequence but nevertheless show multiple
homologous XY gene pairs (gametologs). The latter are generally given the
same gene symbols followed by an X or a Y such as SMCX on proximal Xp
and its equivalent SMCY on Yq. In some cases, however, the Y-chromosome
homologs have degenerated into pseudogenes (with symbols terminating in a
P; see examples in the gene clusters labeled a, b, and c). As a result of
positive selection, the sequence of the male-determinant SRY is now rather
different from its original gene partner on the X chromosome, the SOX3 gene
(highlighted in yellow). [Adapted from Lahn BT & Page DC
(1999) Science 286, 964-967. With permission from the American Association
for the Advancement of Science.]](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-24-320.jpg)



























![RNA genes: A twist in Central Dogma?
• Strachan & Read: Chapter:9, Page-274, [9.3] RNA genes
• Nuclear DNA codes for ~21000 protein coding genes which is XX % of
total genome
• ~85% of nuclear DNA is transcribed, whereas only XX% are protein coding
genes (~21000), thus rest are RNA coding genes (~6000)
• Multi-genic and bidirectional transcription is extensive that explains
more no. of genes in a much smaller encoded region
• ~20000 genes in human (xx billion cells) and also in C. elegans (1000
cells), thus RNA machinery seems to be more important
• ncRNA: Functional Noncoding RNA](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-52-320.jpg)









![Role of ncRNA in epigenetic regulation:
• XIST gene encodes long ncRNA that regulates X-chromosome
inactivation in female mammals [Xq13]
• H19RNA plays a role in repressing transcription of either paternal or
maternal allele of many autosomal regions i.e. imprinting [11p15]
• PEG3RNA plays a role in tumour suppression by activating P53, and is
maternally imprinted [19q13]
• The long mRNA like ncRNA are regulated by genes that produce long
antisense ncRNA transcripts that do not undergo splicing
• HOX gene cluster of 39 genes encode ~231 different long ncRNA](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-62-320.jpg)

![GTG [G banding by Giemsa Trypsin] banded Metaphase cell
Trypsin treatment- digests chromatin protein
increase access to Giemsa stain - Higher
access in
AT rich regions [two Hydrogen bonds] as
compared to
GC rich regions [three Hydrogen bonds]](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-64-320.jpg)



![One-Gene-One-Enzyme,
Pseudogenes
& Common Ancestry
The following animation is intended to show:
1. The one-gene-one-enzyme hypothesis
2. How a mutation in one gene (probably in some
early pre-primate) prevented the production of
Vitamin C, explaining why all primates today
require Vitamin C in their diets (not so with
other mammals).
3. The GULO pseudogene evidence for the
common ancestry of primates [Gene coding for
enzyme L-gulonolactone oxidase]
68](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-68-320.jpg)








![• Archibald Garrod: Inborn errors of metabolism, example of
Alkaptonuria [his definition, one mutant gene one metabolic block]
• George Beadle & Boris Ephrussi on Drossophila (1930s); Beadle &
Edward Tatum on Neurospora Crassa: One gene one enzyme using
X-ray irradiation of spores and growth on complete medium, Nobel
prize in 1958](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-77-320.jpg)







![Bacteriophage øX174: Overlapping Genes &
Genes within genes
•5386 nucleotide- 11 genes- 2300 amino
acids
•5386/3 1795 amino acids expected [500
lesser than observed]](https://image.slidesharecdn.com/2unitgenomeorganization-230413152858-879abdef/85/2UnitGenomeOrganization-pptx-85-320.jpg)