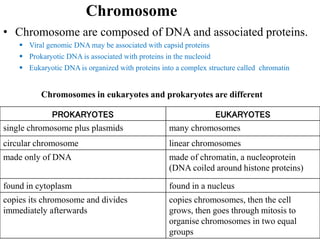
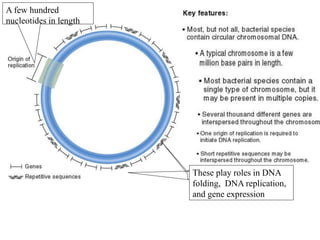
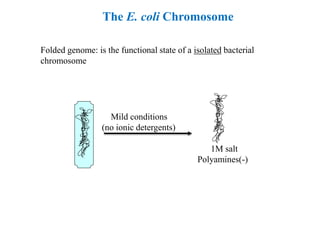
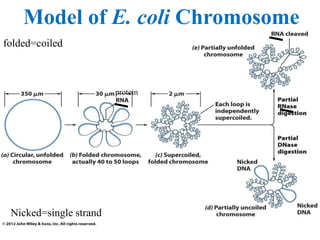
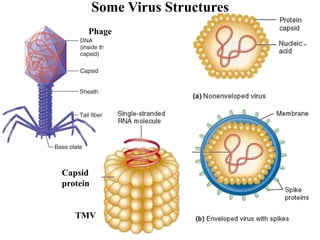
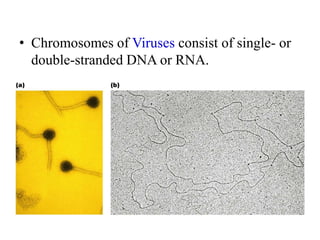
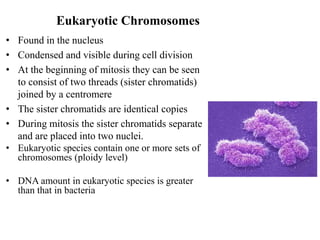
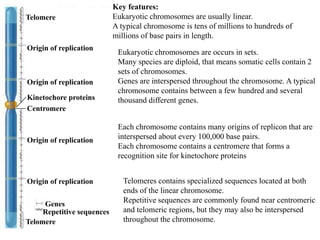
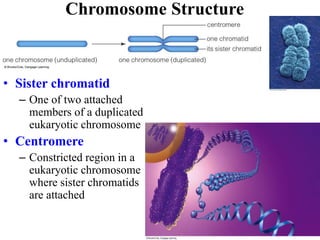
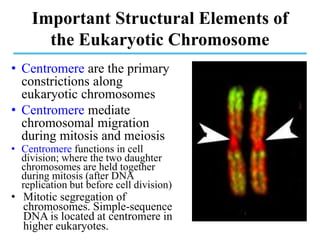
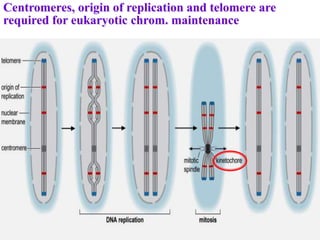
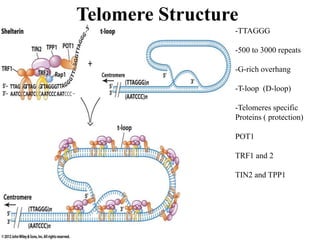
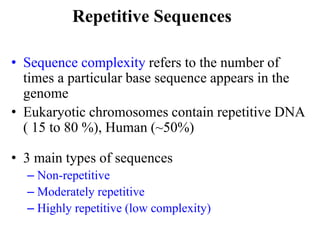
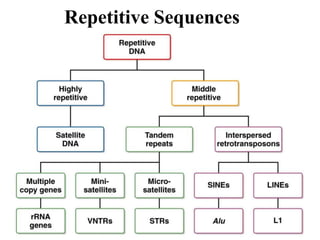
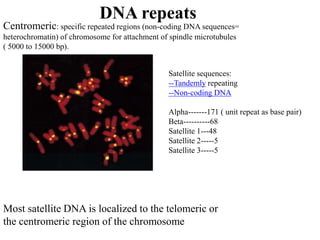
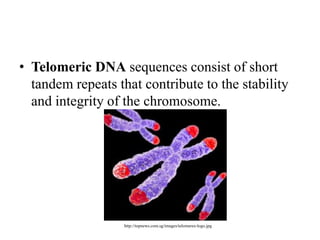
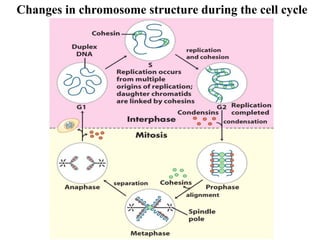
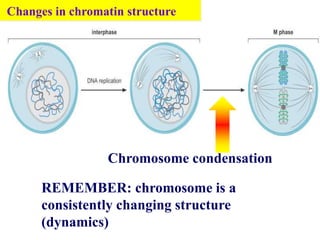
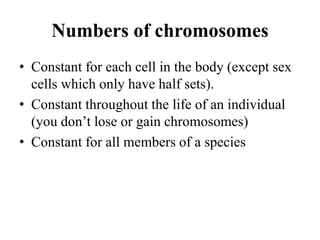
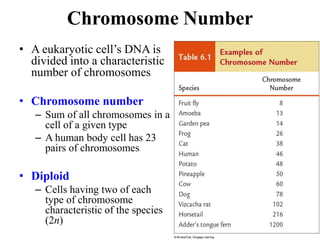
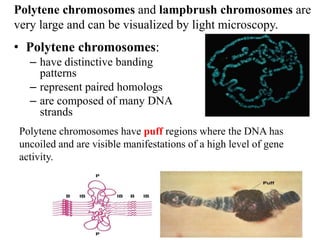
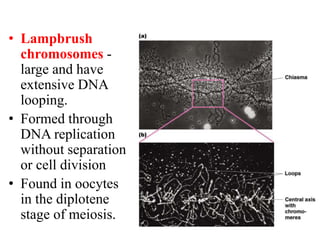
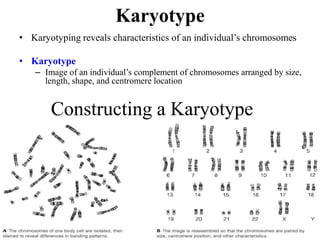
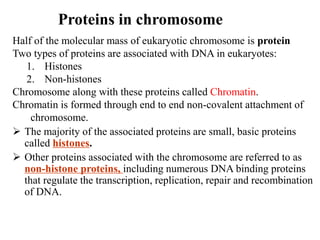
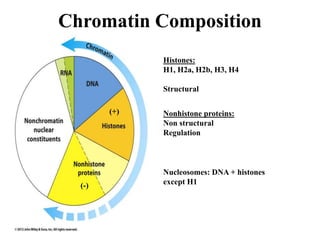
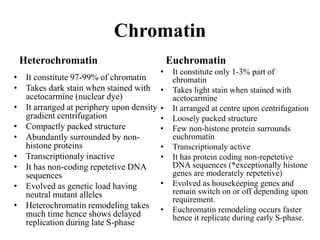
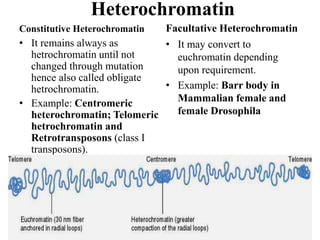
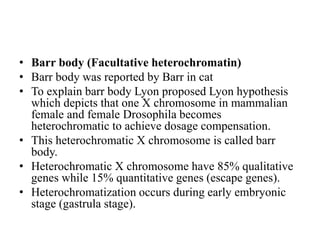
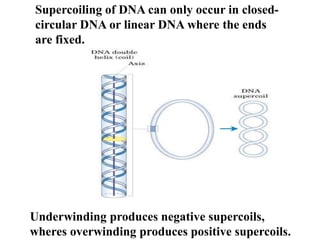
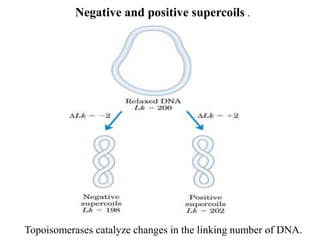
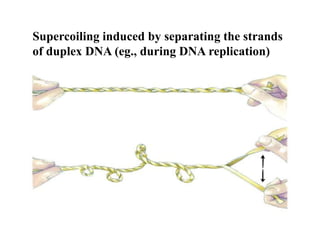
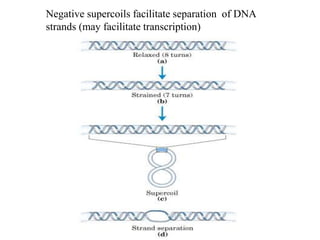
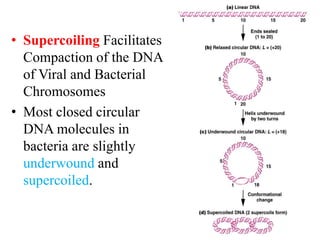
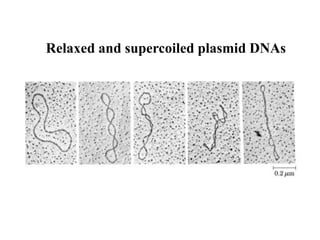
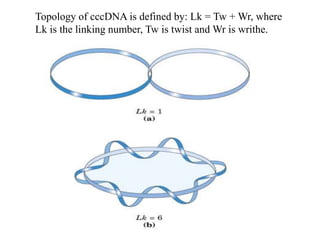
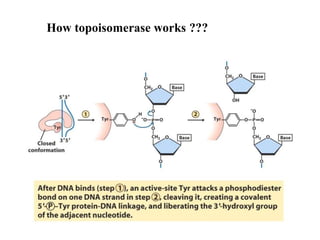
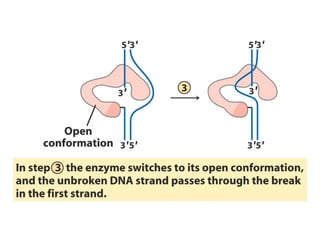
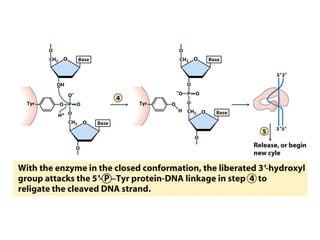
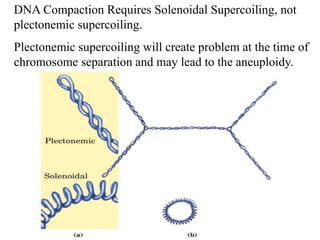
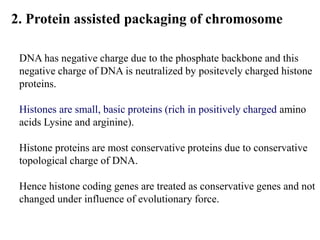
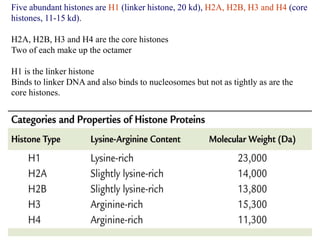
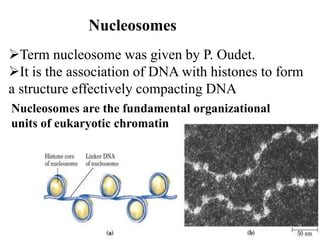
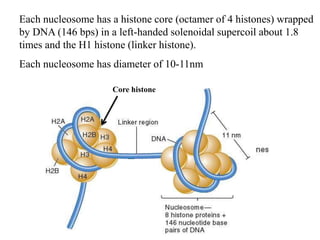
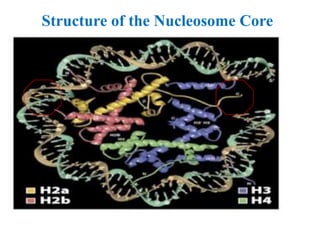
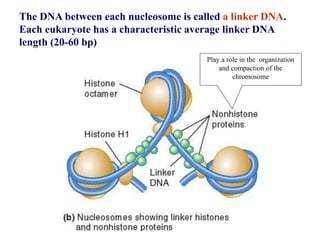
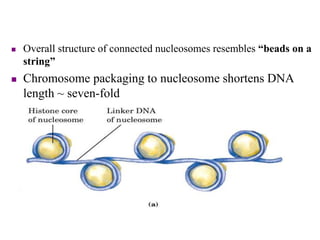
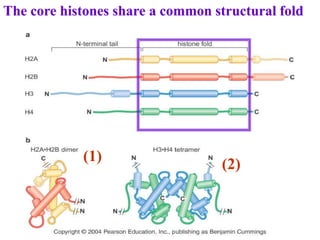
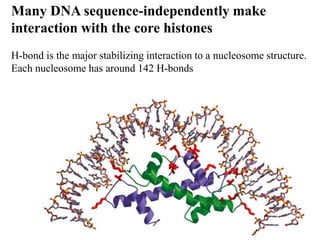
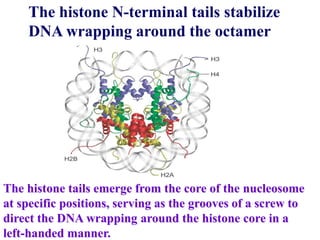
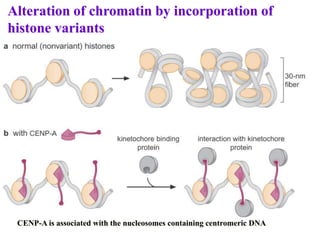
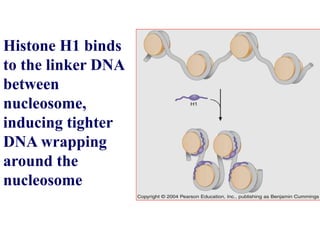
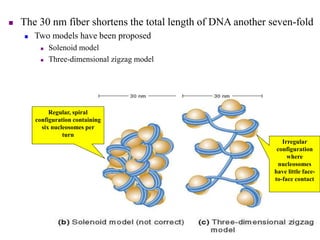
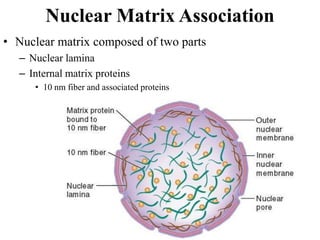
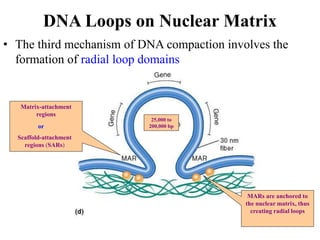
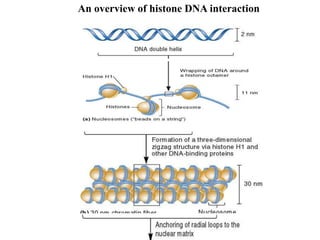
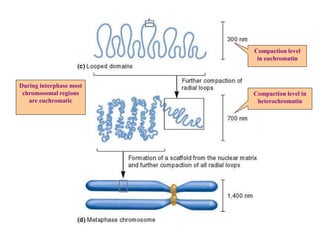
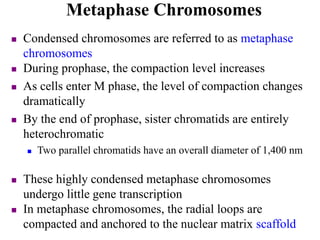
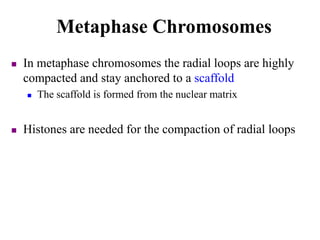
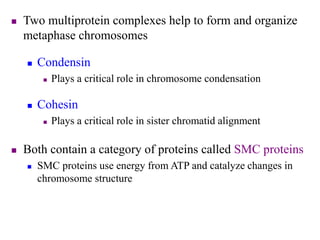
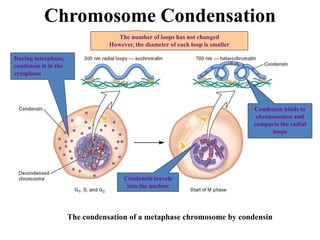
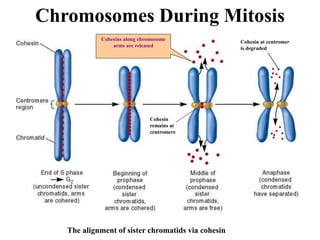
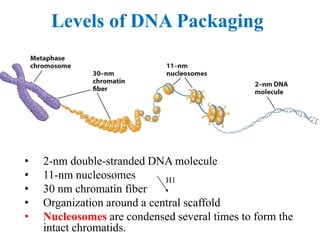
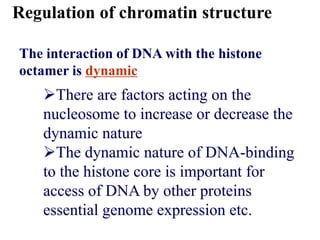
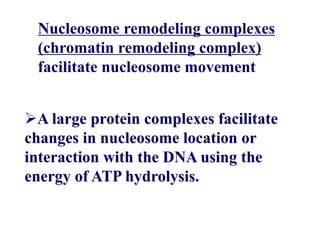
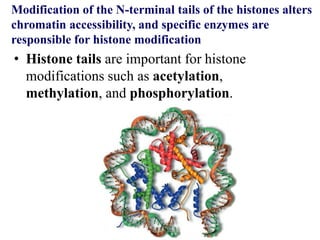
Chromosomes are structures that package and organize DNA and associated proteins. In eukaryotes, DNA is wrapped around histone proteins to form chromatin, which condenses into linear or circular chromosomes. Key features of eukaryotic chromosomes include centromeres, telomeres, and repetitive sequences. Chromosomes are compacted through DNA supercoiling and packaging into nucleosomes. The structure and packaging of chromosomes allows for efficient storage and regulation of the genetic material.