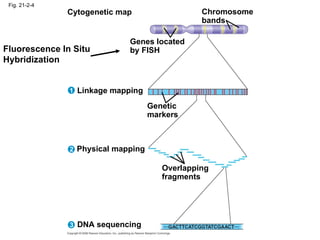
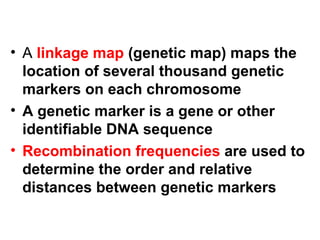
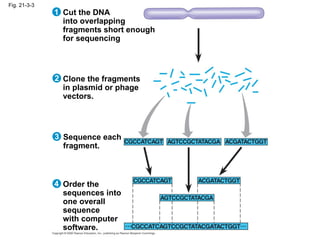
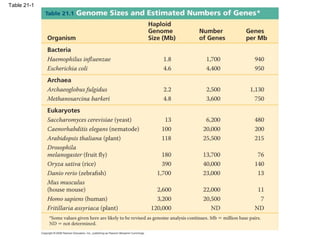
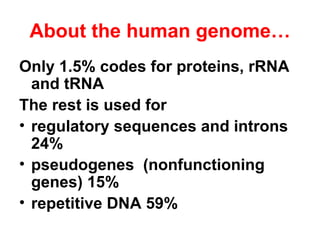
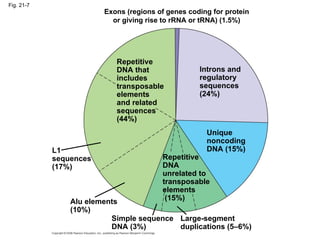
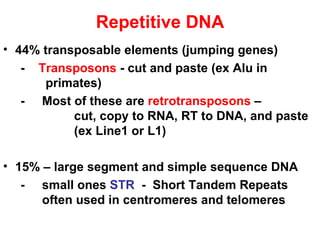
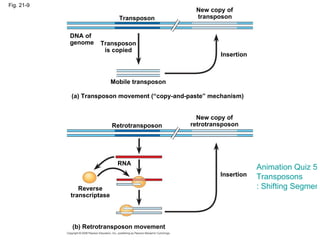
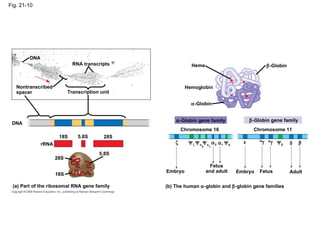
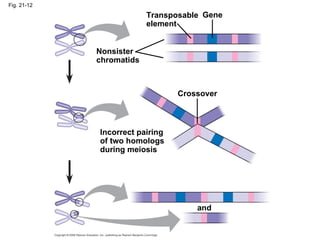
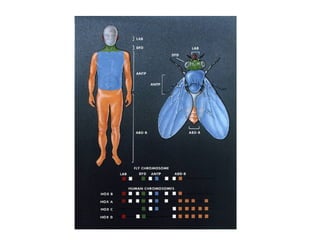
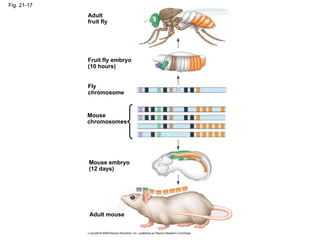
The document summarizes key aspects of genomics and the human genome project. It discusses that the human genome project mapped the human genome through linkage mapping, physical mapping, and DNA sequencing. It was completed in 2003 and found that humans have around 20,000 genes and repetitive non-coding DNA makes up around half the genome. Transposons and retrotransposons are types of repetitive DNA that can move locations within genomes.