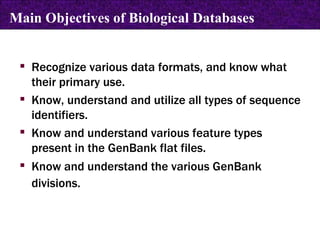
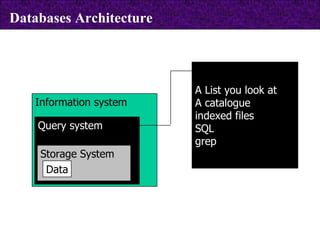
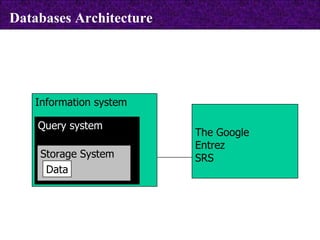
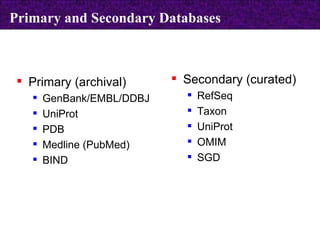
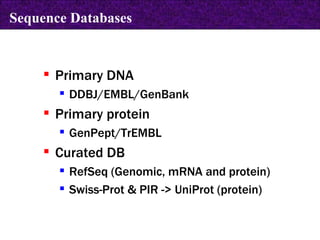
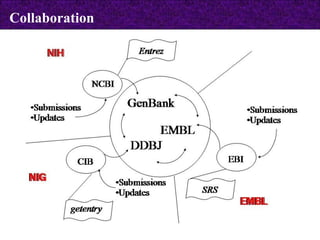
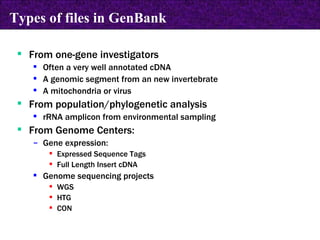
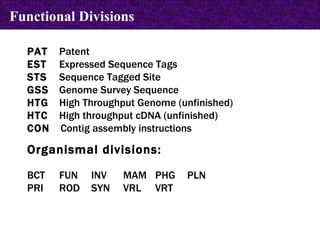
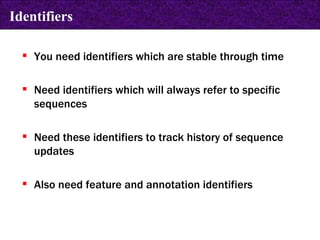
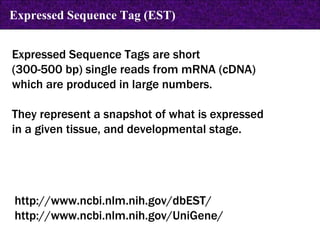
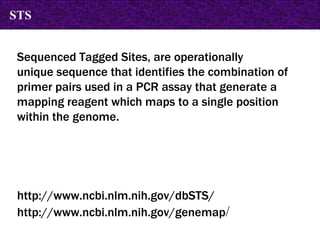
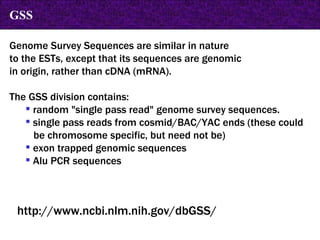
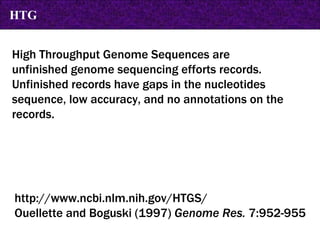
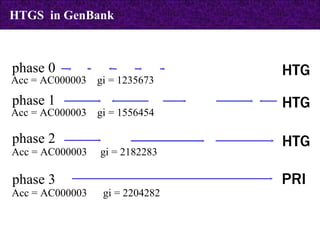
The document provides information about biological databases and sequence identifiers. It discusses the main objectives of biological databases which include information systems, query systems, storage systems and data. It describes primary databases like GenBank, EMBL, DDBJ, UniProt and PDB as well as secondary curated databases like RefSeq, Taxon and OMIM. It also explains different types of sequence identifiers used in databases like LOCUS, ACCESSION, VERSION, gi numbers and protein identifiers.
![S.Prasanth Kumar, Bioinformatician Biological Databases Bioinformatics and Biostatistics S.Prasanth Kumar, Bioinformatician S.Prasanth Kumar Dept. of Bioinformatics Applied Botany Centre (ABC) Gujarat University, Ahmedabad, INDIA www.facebook.com/Prasanth Sivakumar FOLLOW ME ON ACCESS MY RESOURCES IN SLIDESHARE prasanthperceptron CONTACT ME [email_address]](https://image.slidesharecdn.com/biologicaldatabases-110324013536-phpapp02/75/Biological-databases-1-2048.jpg)

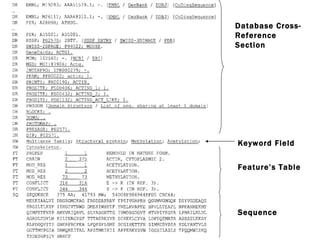
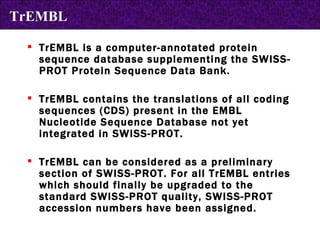
![SWISS-PROT an annotated universal sequence database, TrEMBL an automatically generated sequence database with repository character, which supplements SWISS-PROT. SWISS-PROT [http://www.expasy.org/sprot/] a curated protein sequence database which provides a high level of annotation Types of Annotations: Description of a protein's function, its domain structure, PTMs, conflicts between literature references and variants. It also provides a minimal level of redundancy , a high level of integration with other bio molecular databases , and an extensive external documentation. (Created in 1986:Main Host-ExPaSy) Protein sequence databases](https://image.slidesharecdn.com/biologicaldatabases-110324013536-phpapp02/85/Biological-databases-16-320.jpg)

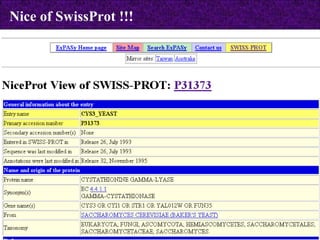



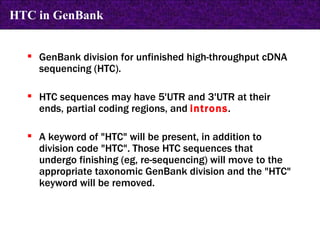


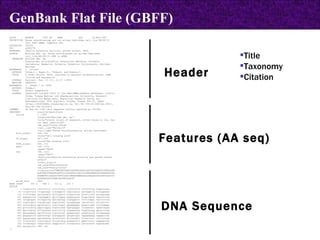
![LOCUS AH007743 7832 bp DNA CON 26-MAY-1999 DEFINITION Gallus gallus ornithine transcarbamylase (OTC) gene, complete cds. ACCESSION AH007743 VERSION AH007743.1 GI:4927367 KEYWORDS . SOURCE chicken. ORGANISM Gallus gallus Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Archosauria; Aves; Neognathae; Galliformes; Phasianidae; Phasianinae; Gallus. [....] FEATURES Location/Qualifiers source 1..7832 /organism="Gallus gallus" /db_xref="taxon:9031" /chromosome="1" CONTIG join(AF065630.1:1..1903,gap(),AF065631.1:1..435,gap(), AF065632.1:1..509,gap(),AF065633.1:1..722,gap(),AF065634.1:1..707, gap(),AF065635.1:1..836,gap(),AF065636.1:1..1614,gap(), AF065637.1:1..605,gap(),AF065638.1:1..501) // CON in GenBank](https://image.slidesharecdn.com/biologicaldatabases-110324013536-phpapp02/85/Biological-databases-39-320.jpg)






