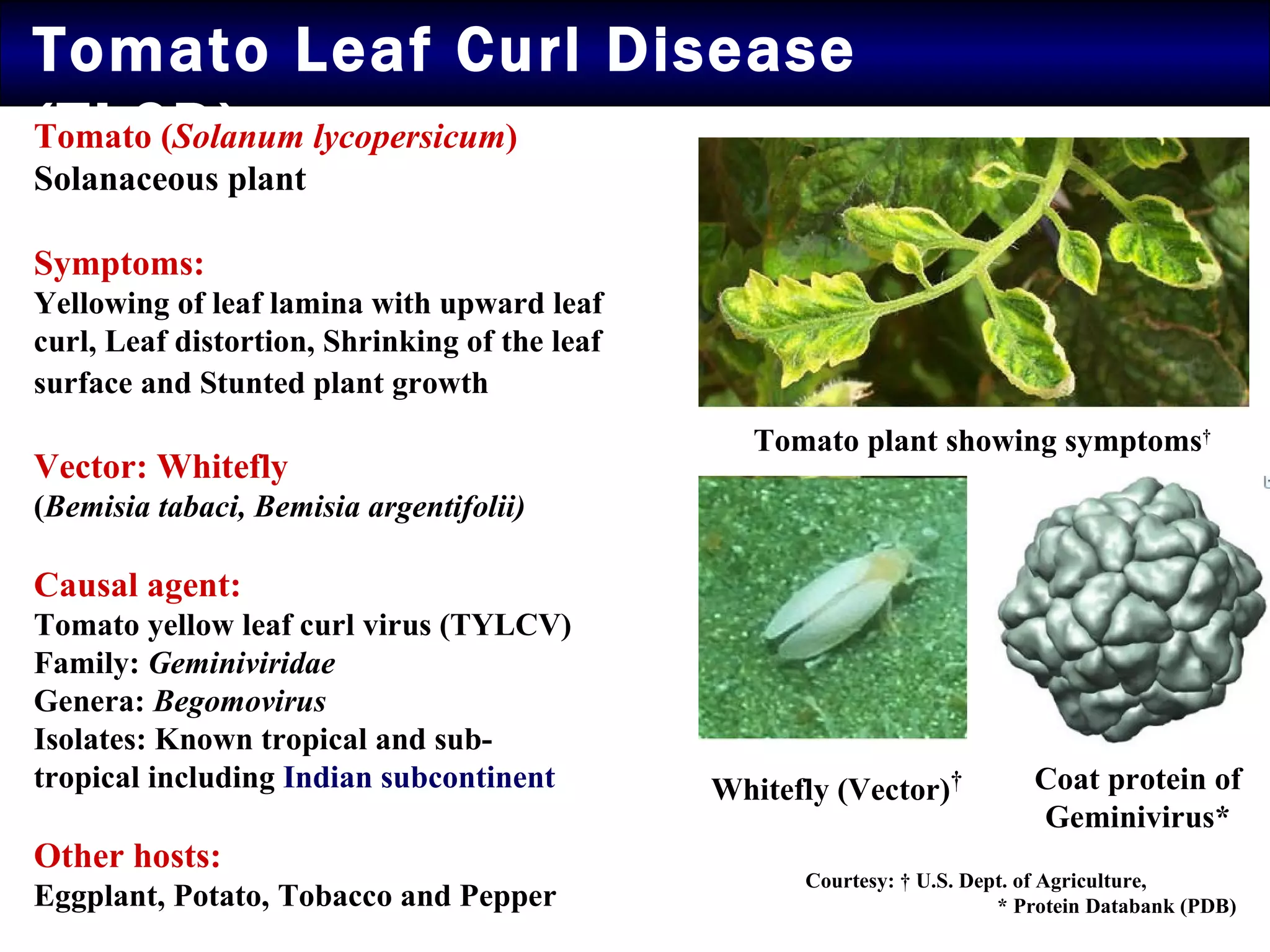
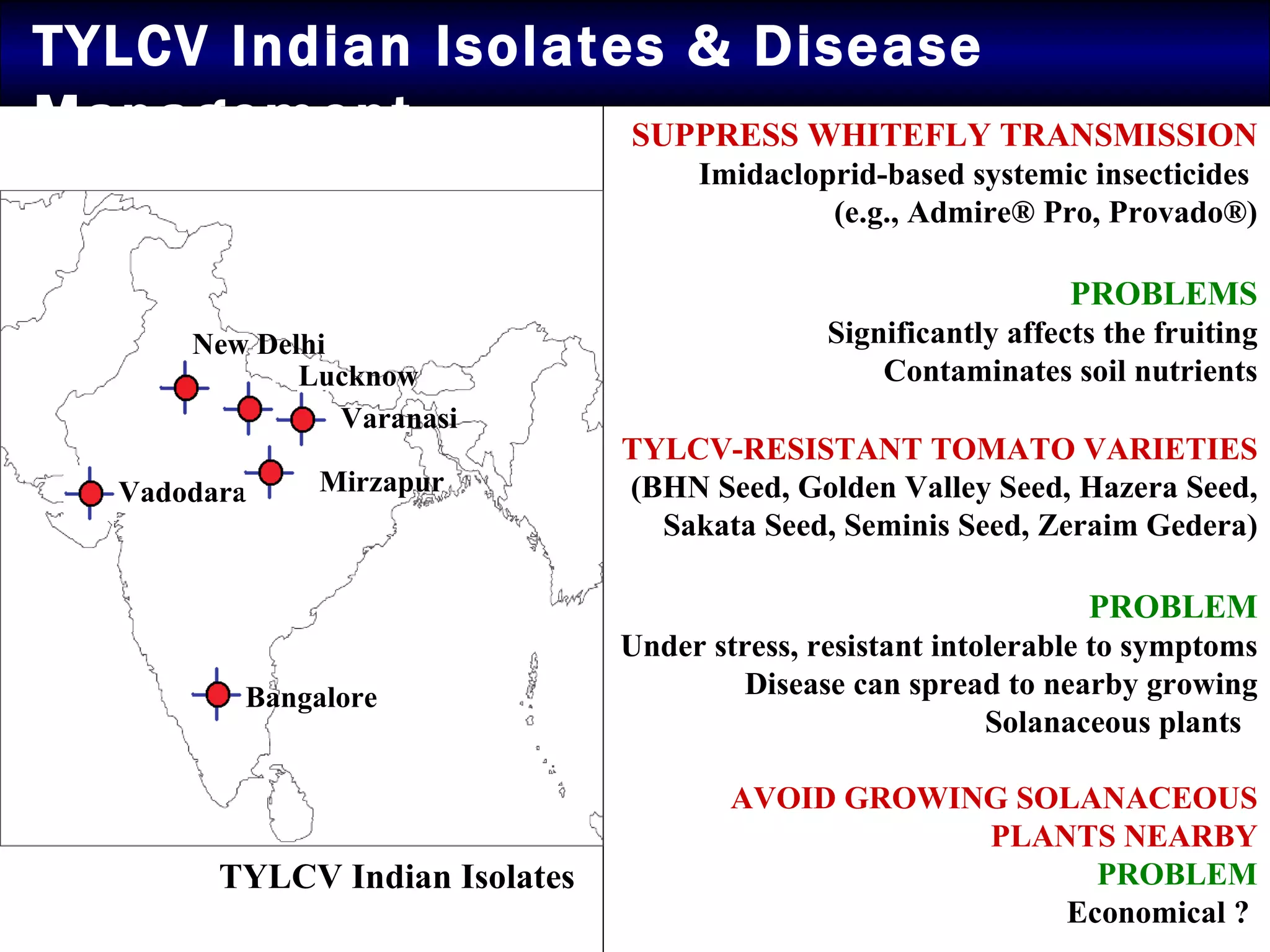
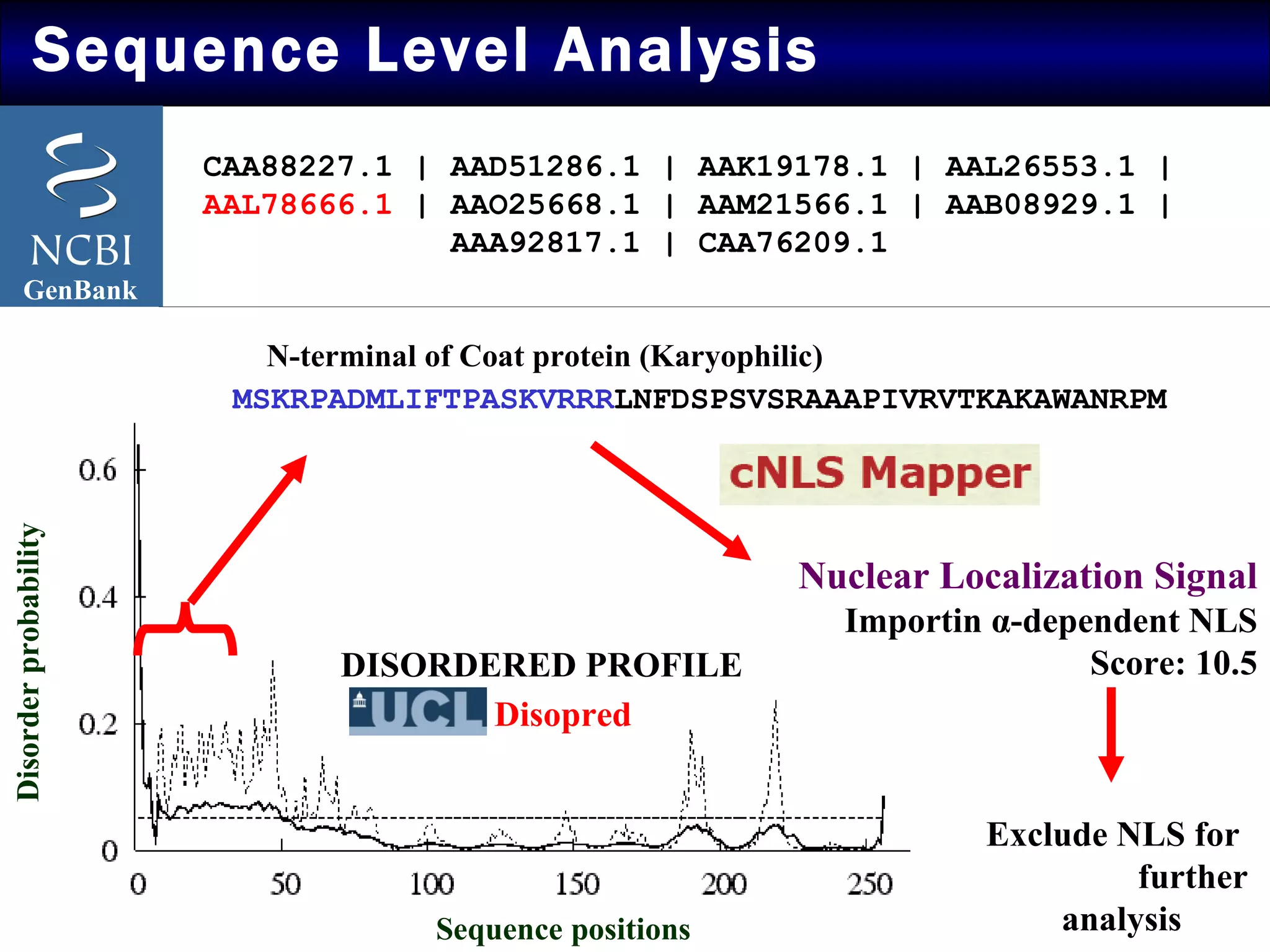
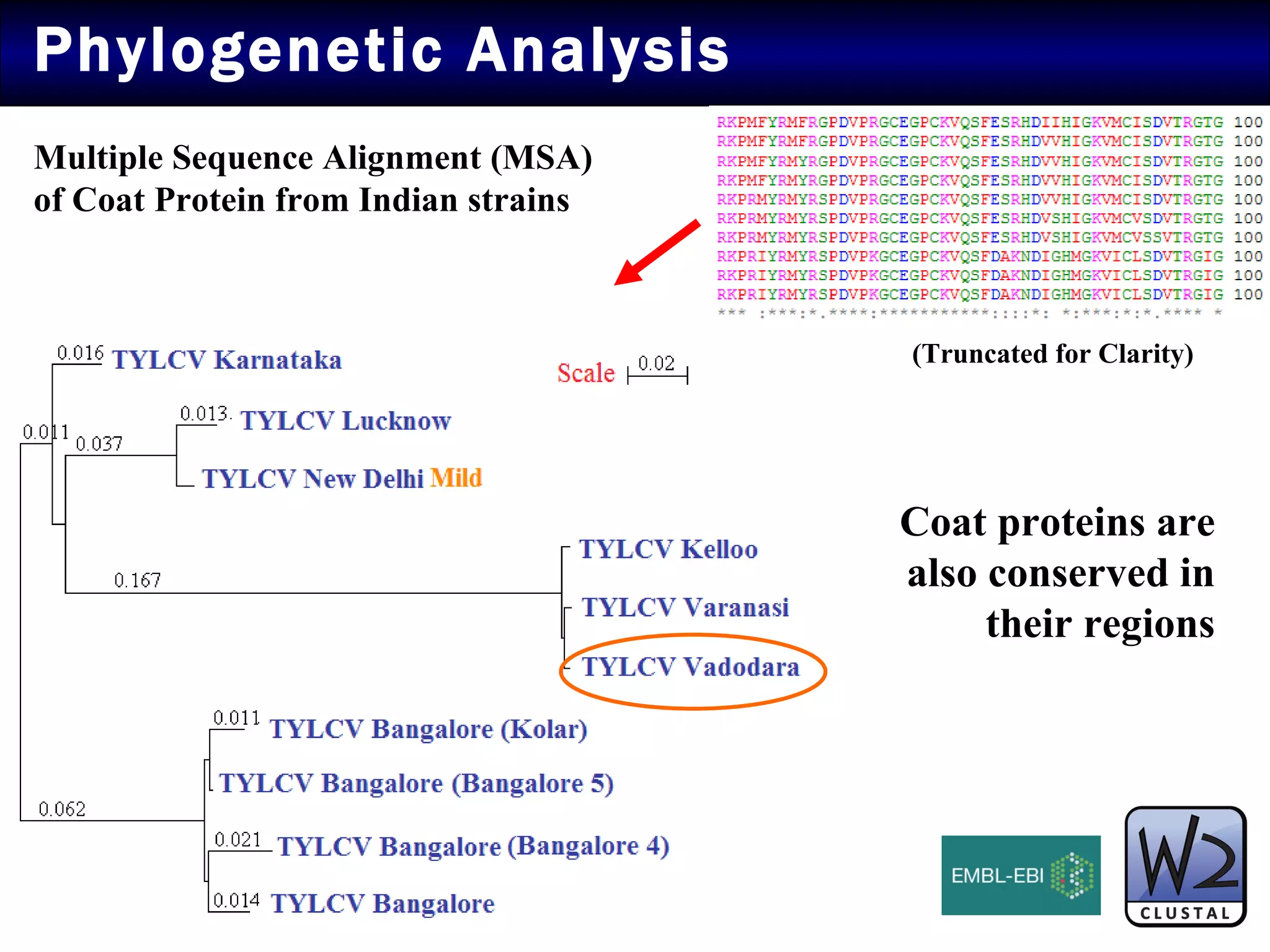
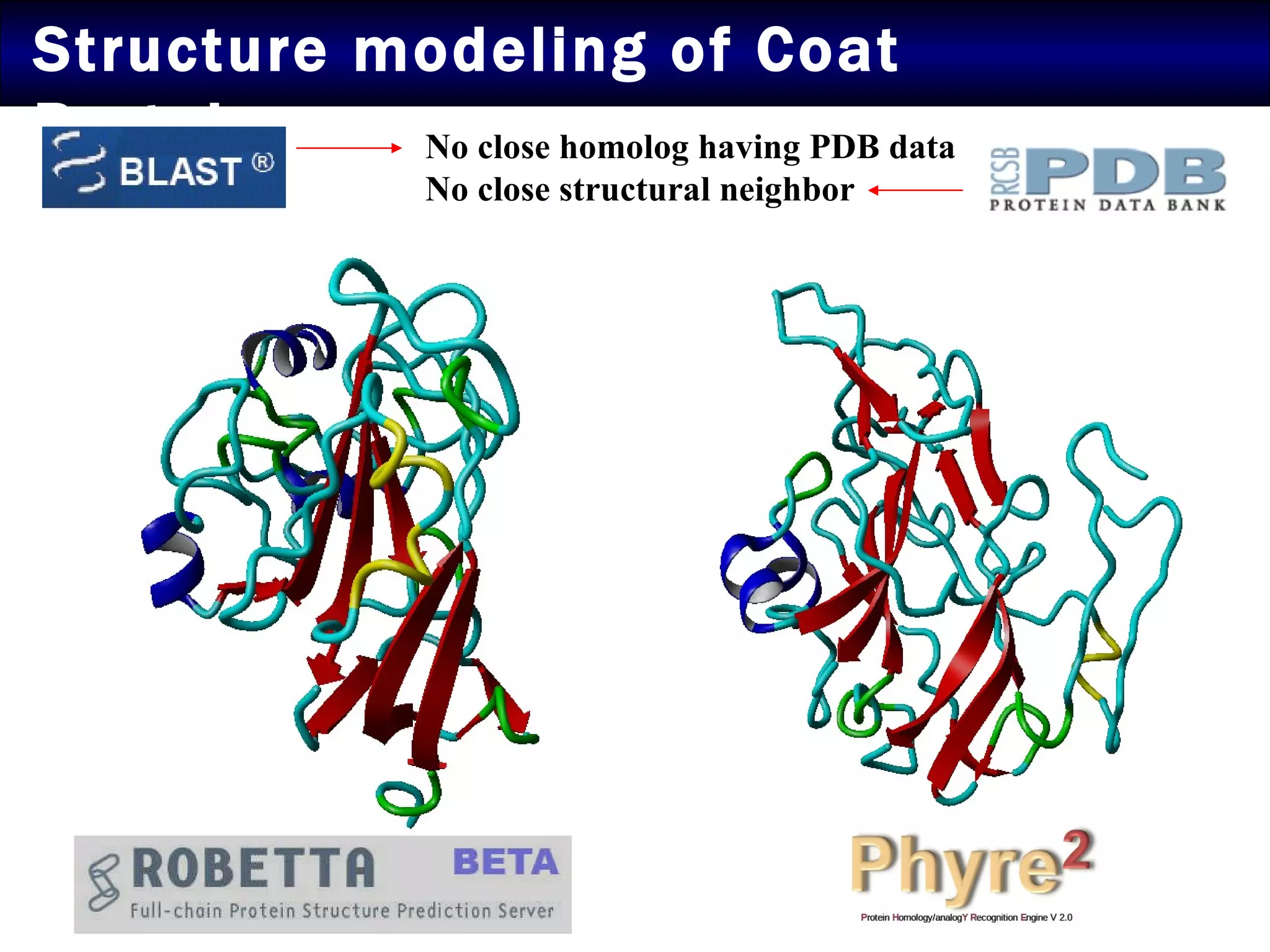
The document summarizes research on the coat protein of the Tomato yellow leaf curl viral (TYLCV) disease, which causes significant damage to tomato crops in India. Key findings include:
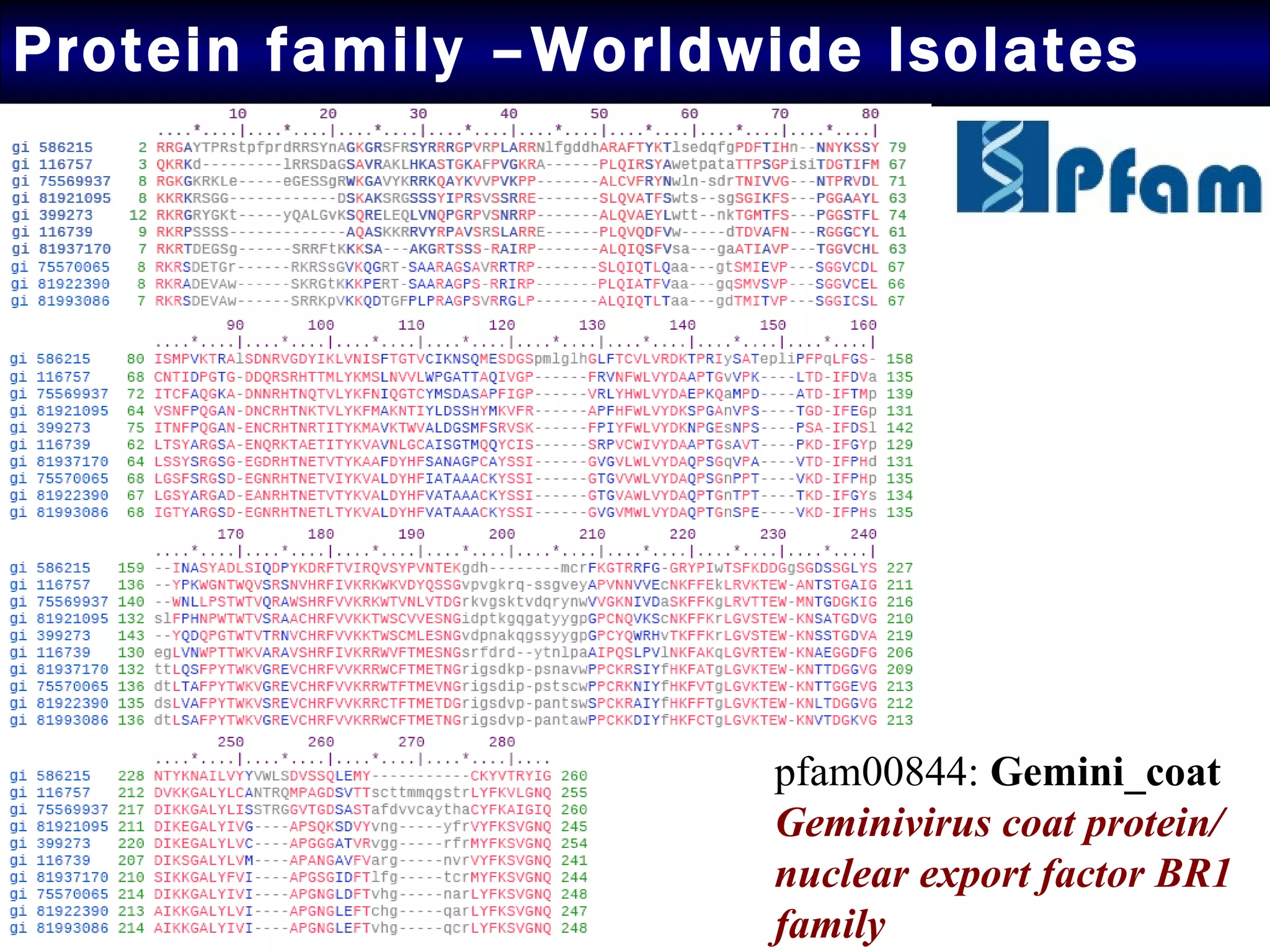
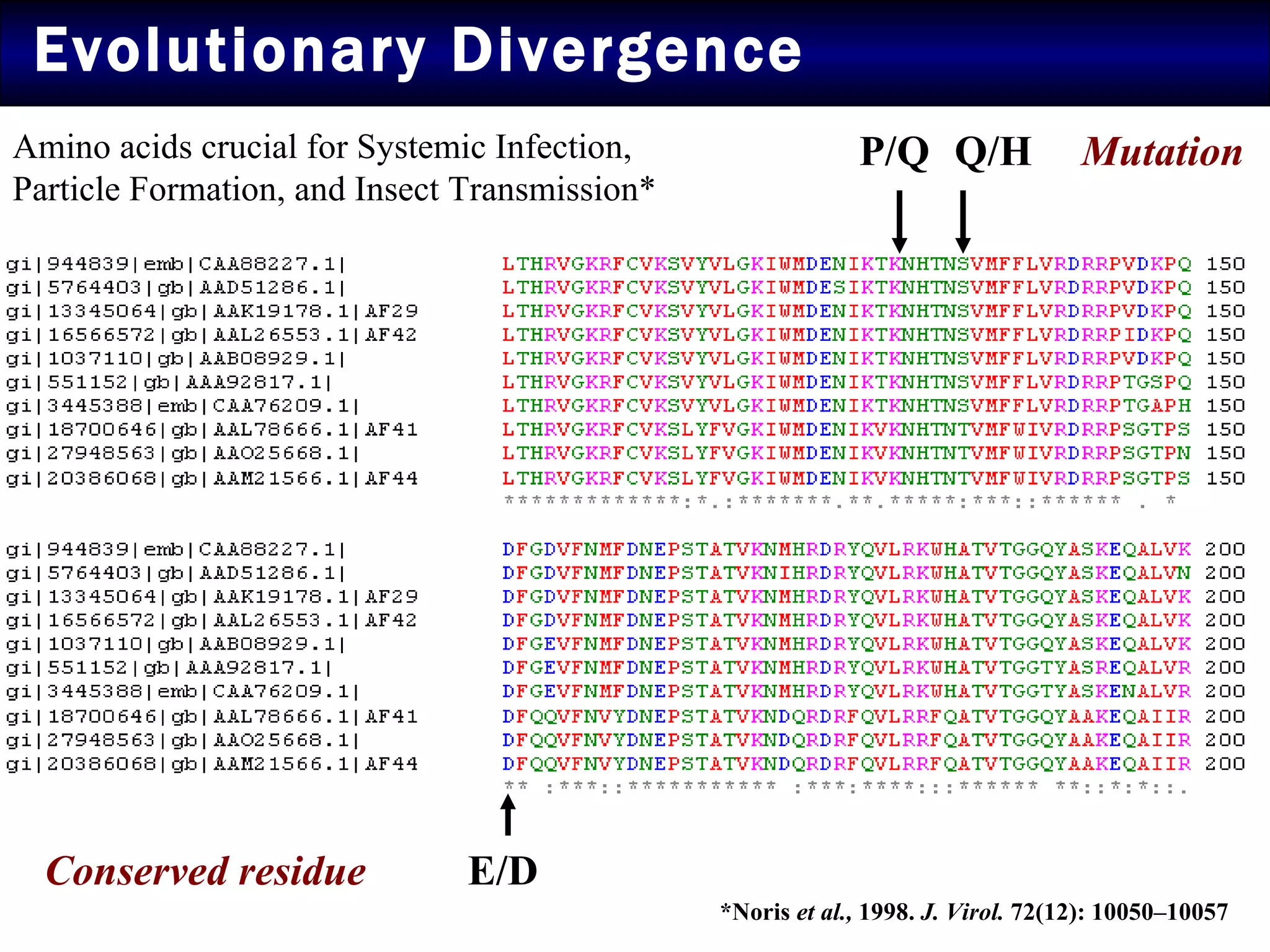
1) Sequence analysis found mutations in the coat protein that provide insights into the evolutionary divergence of Indian virus isolates and their ability to systematically infect plants.
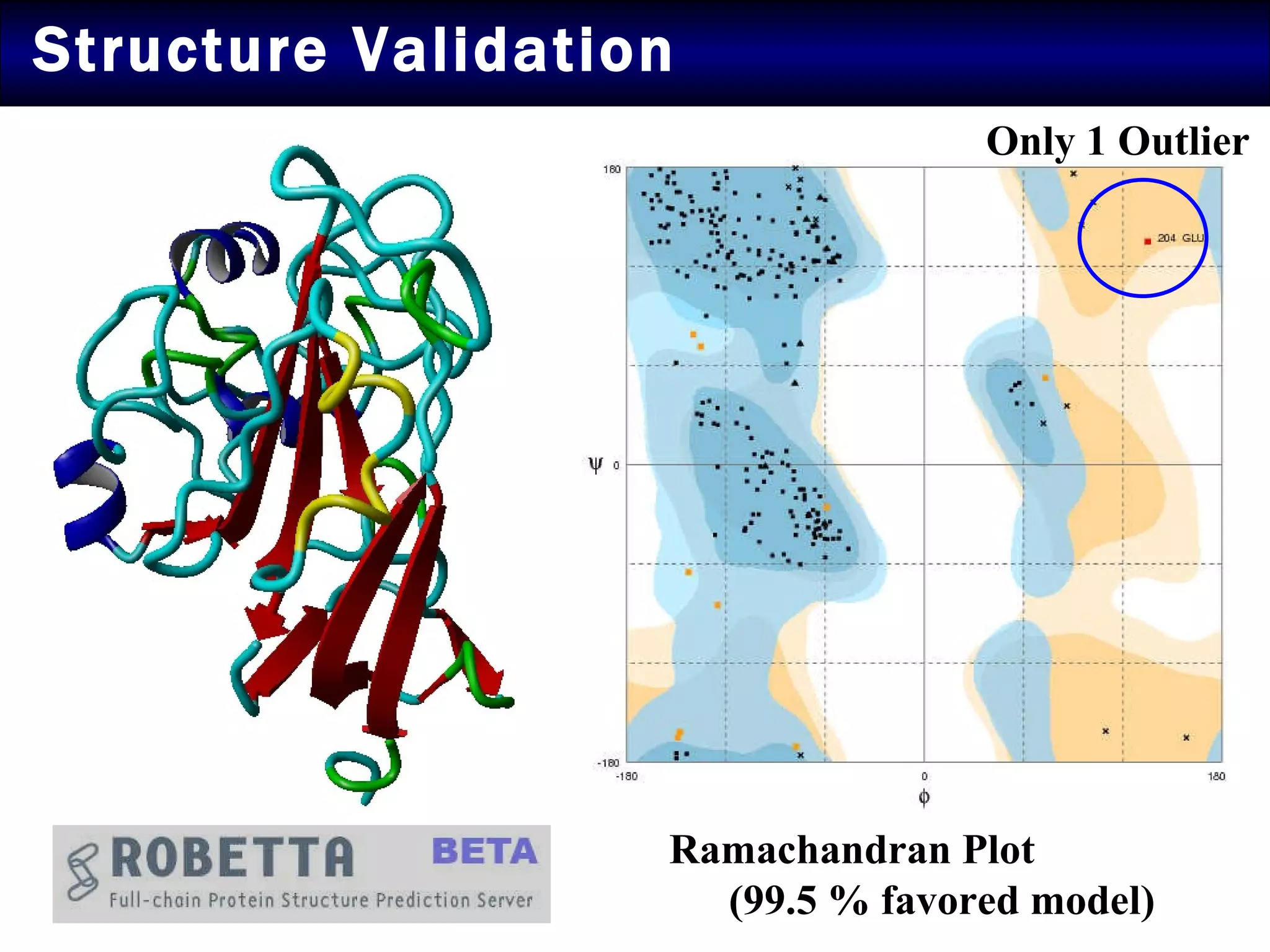
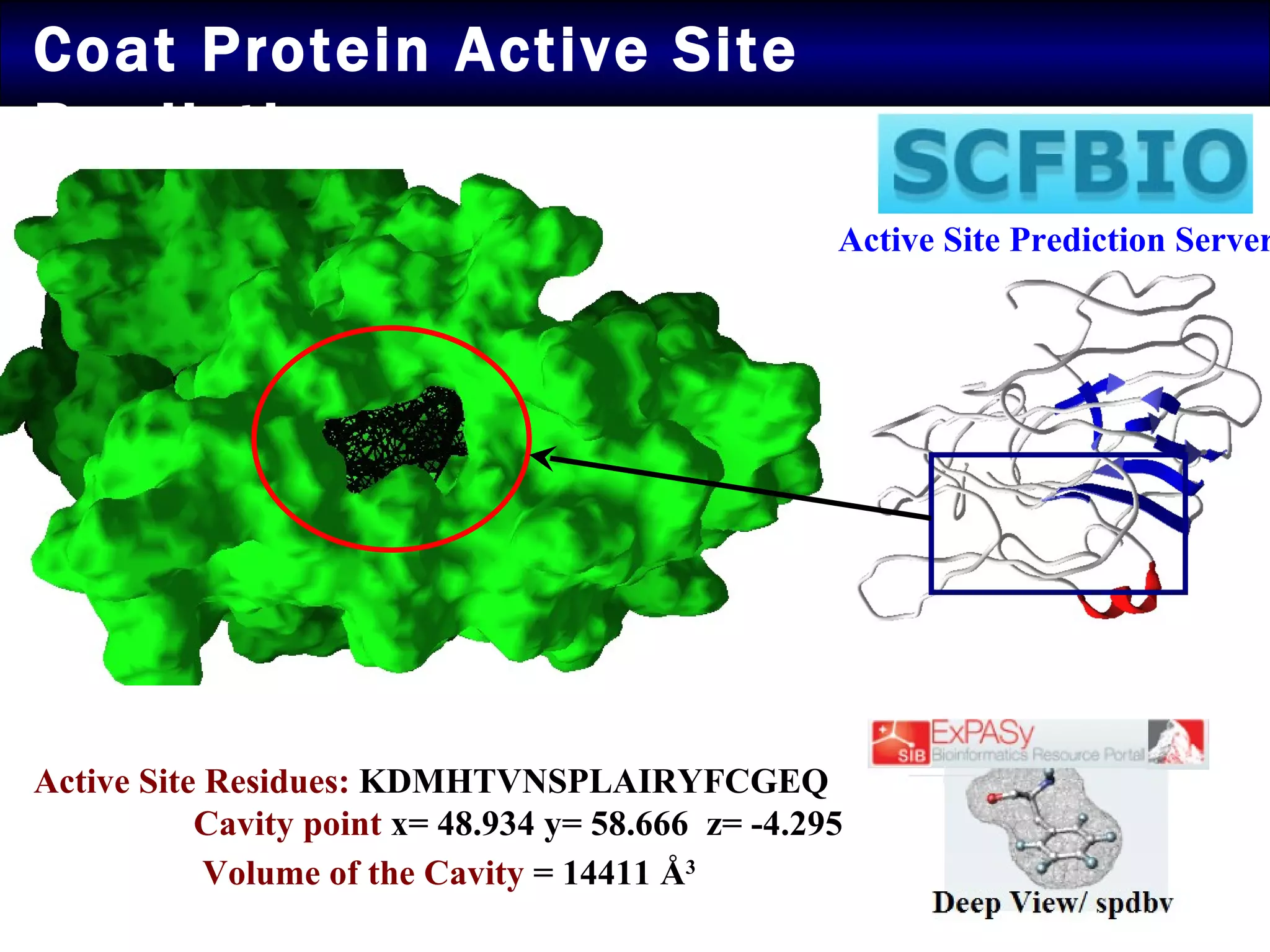
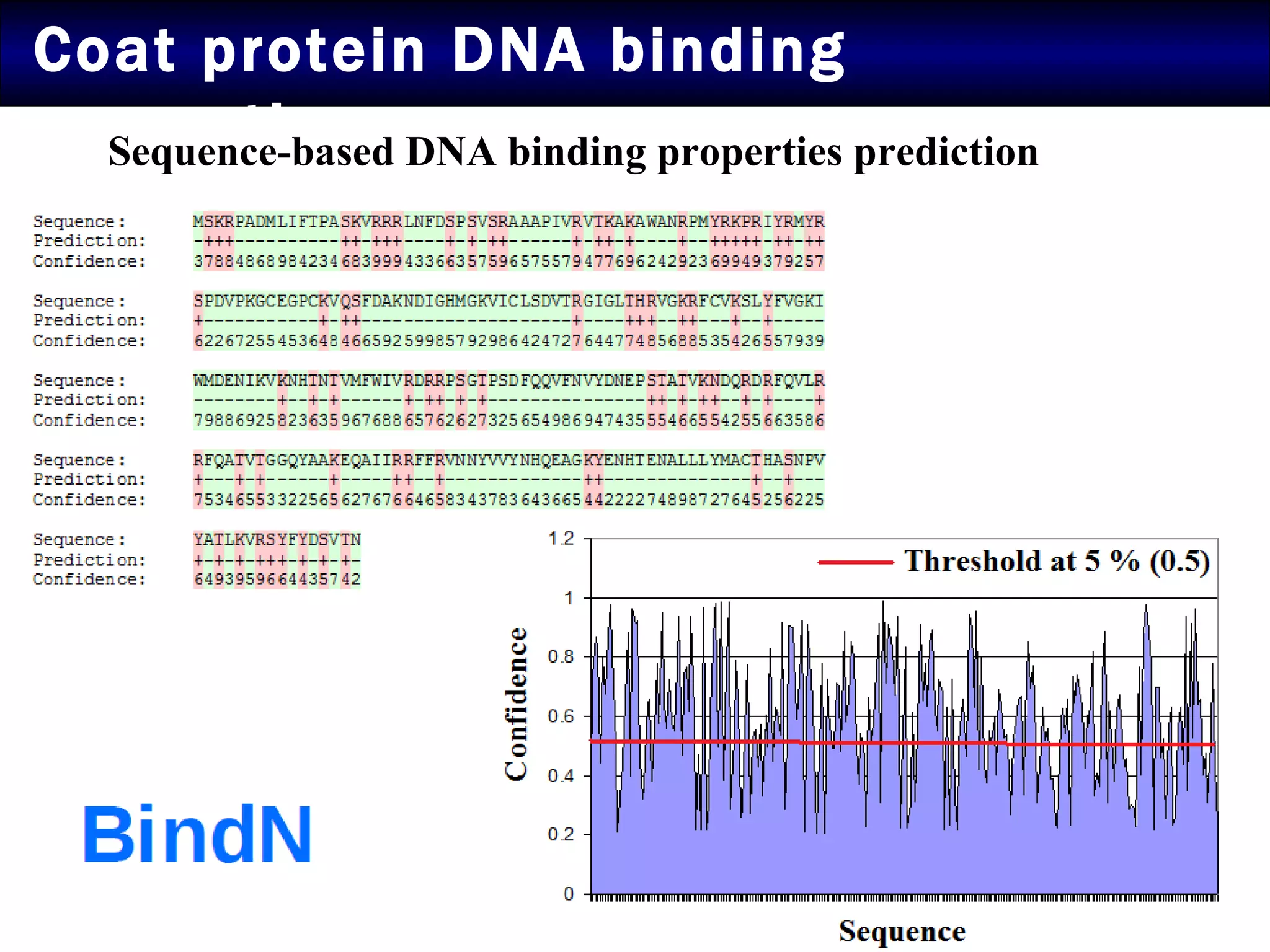
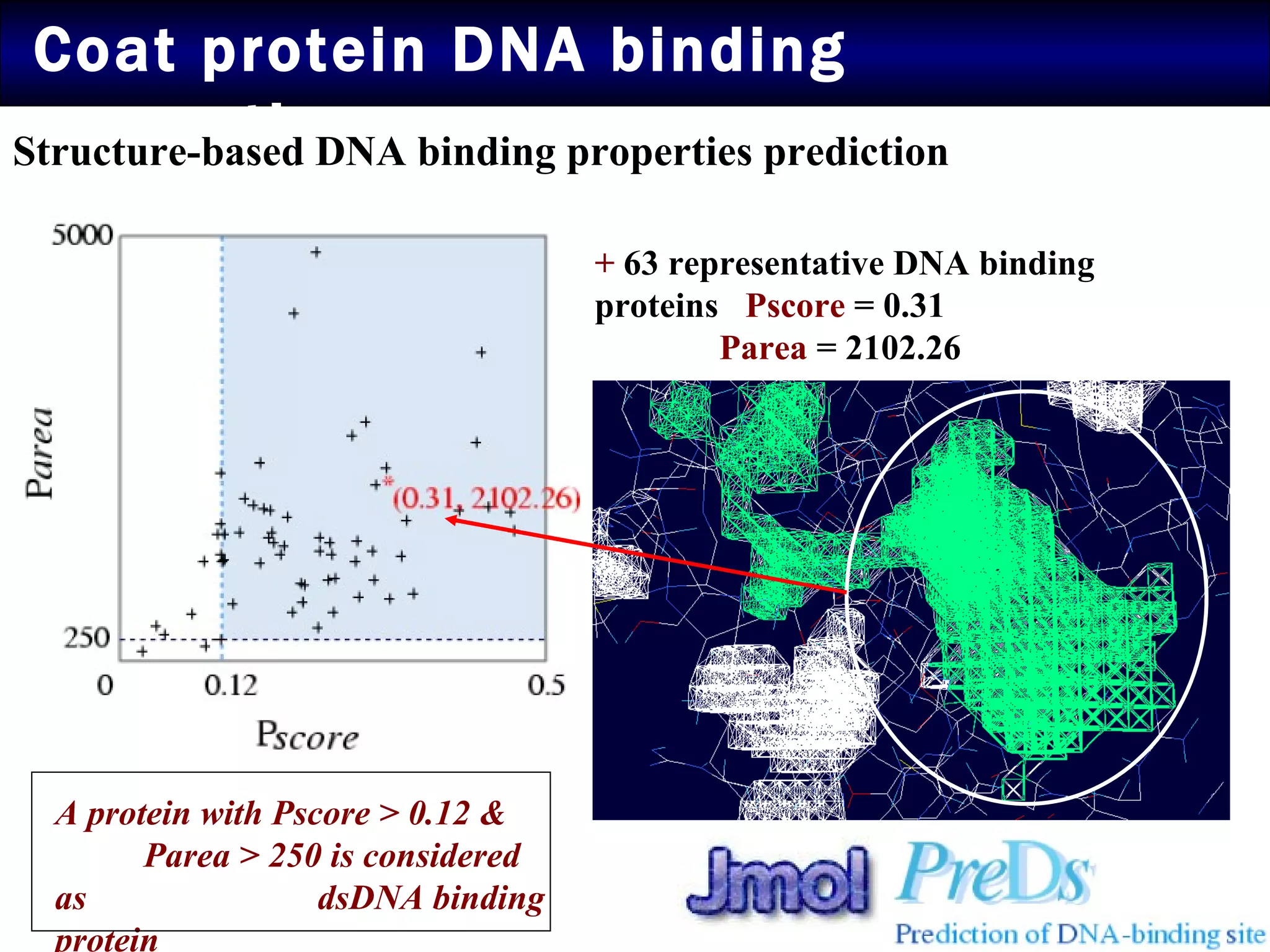
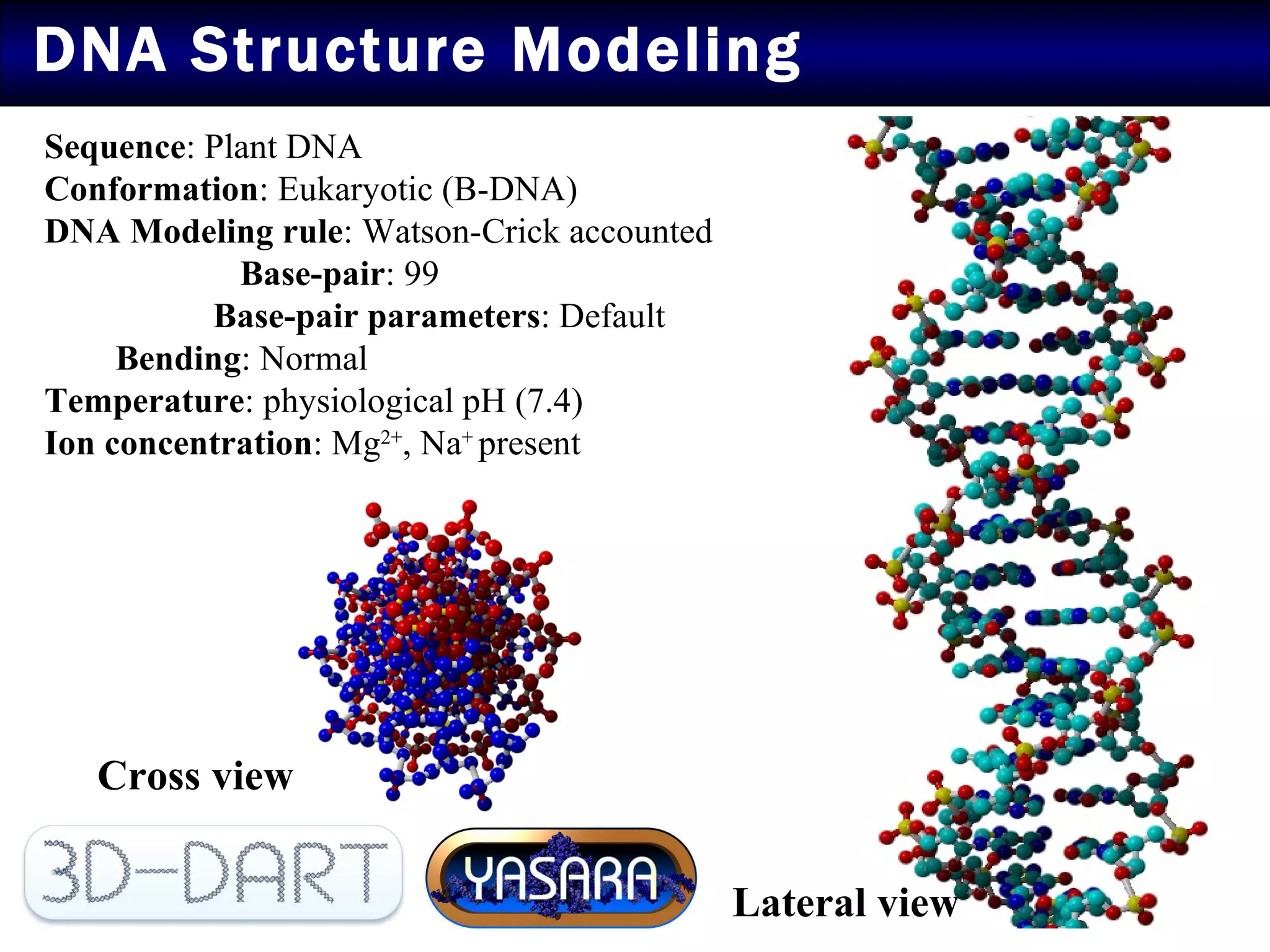
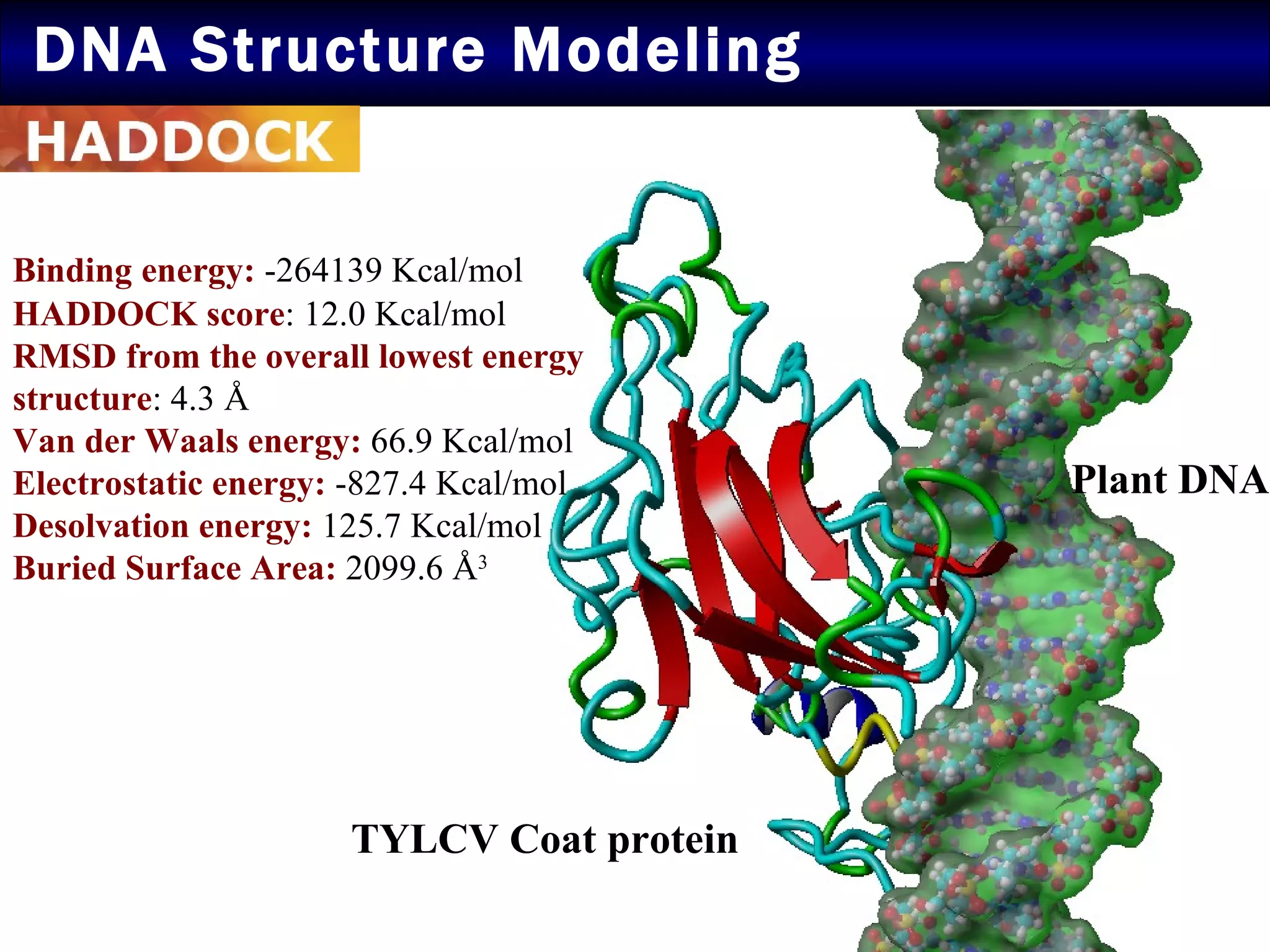
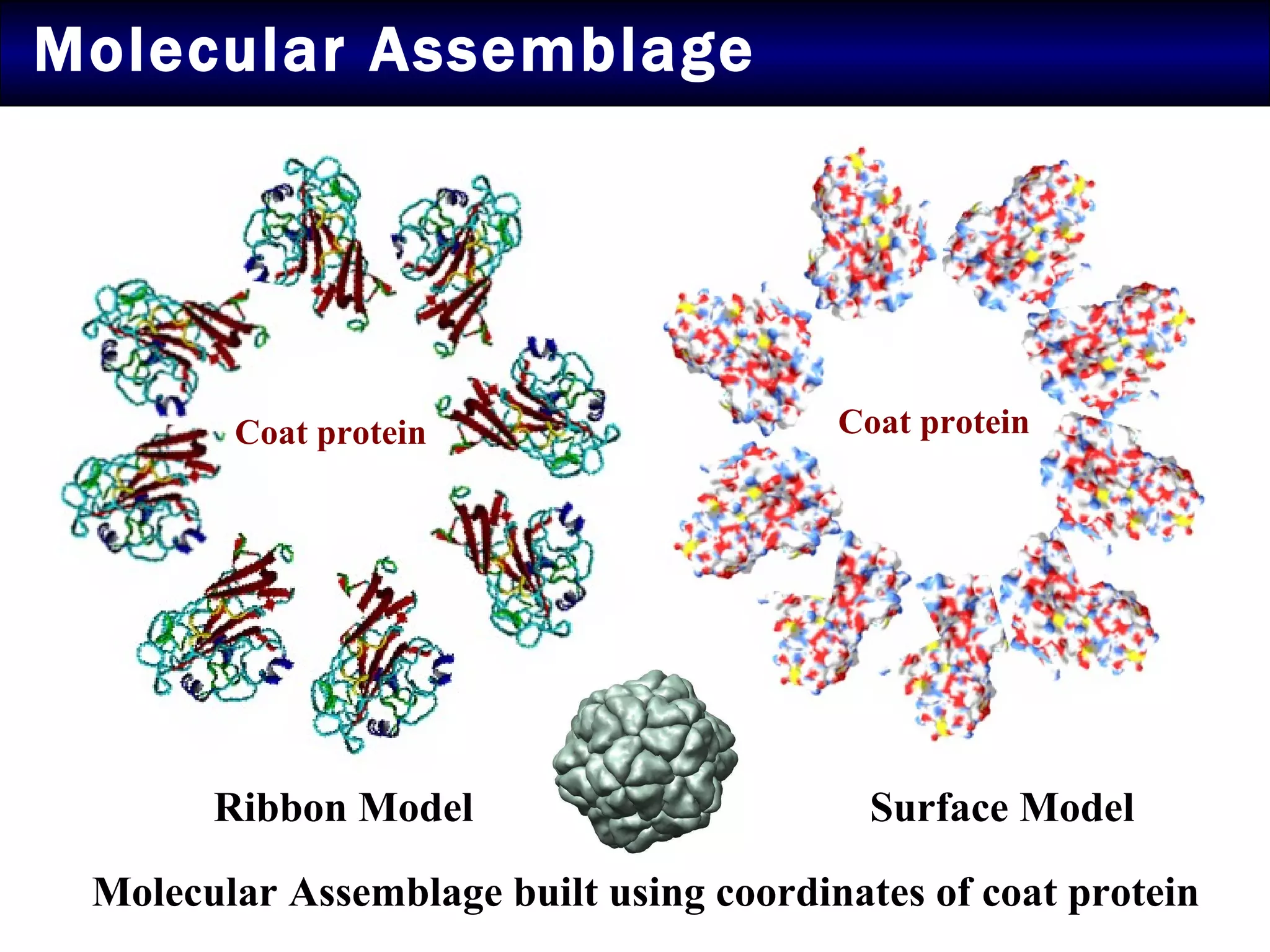
2) Structural modeling predicted the coat protein binds double-stranded DNA through interactions facilitated by surface loops and neutral patches on the protein.
3) Docking simulations showed the coat protein binds plant DNA through electrostatic and van der Waals interactions, helping the virus infect tomato and other plants.