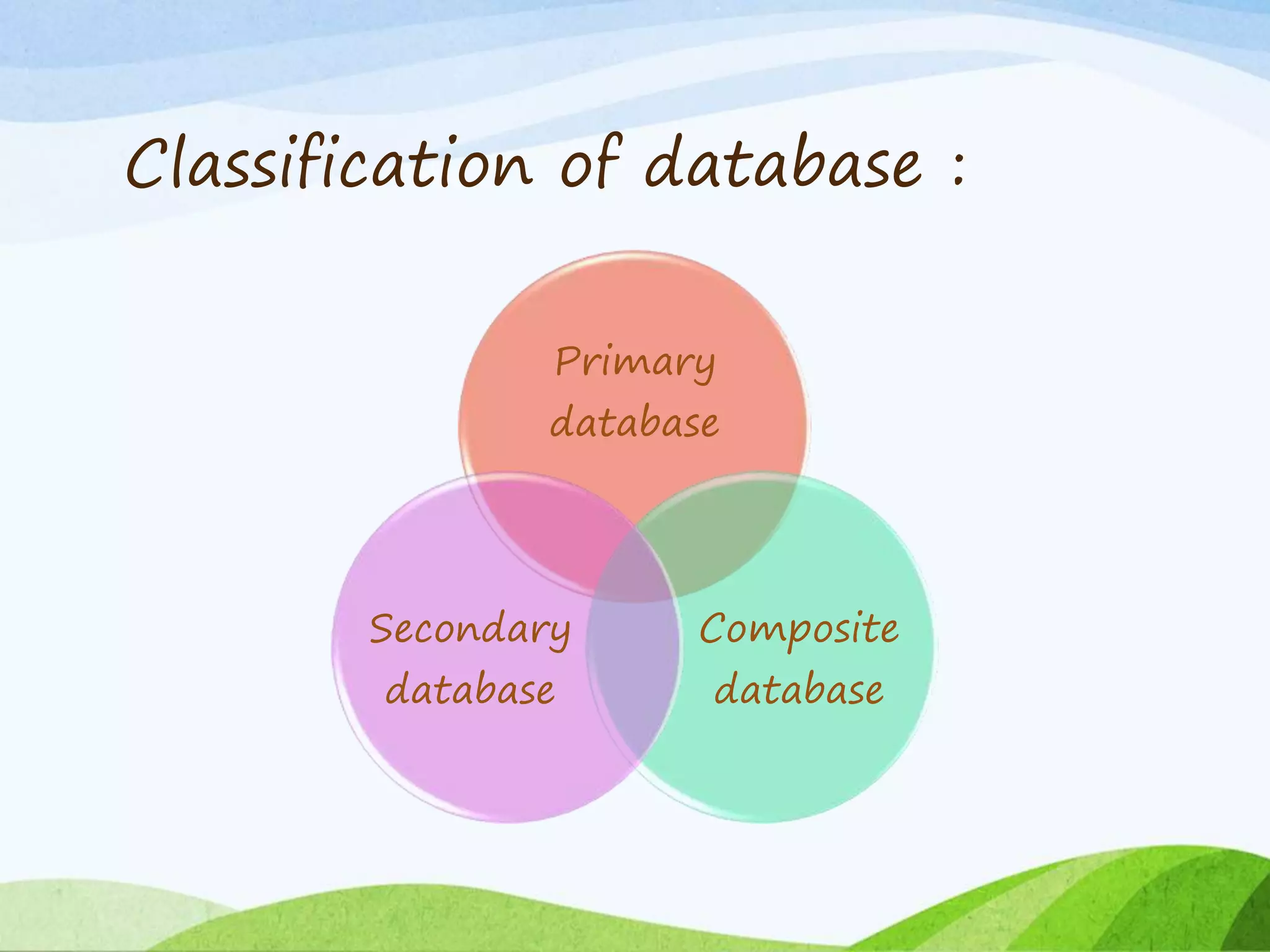
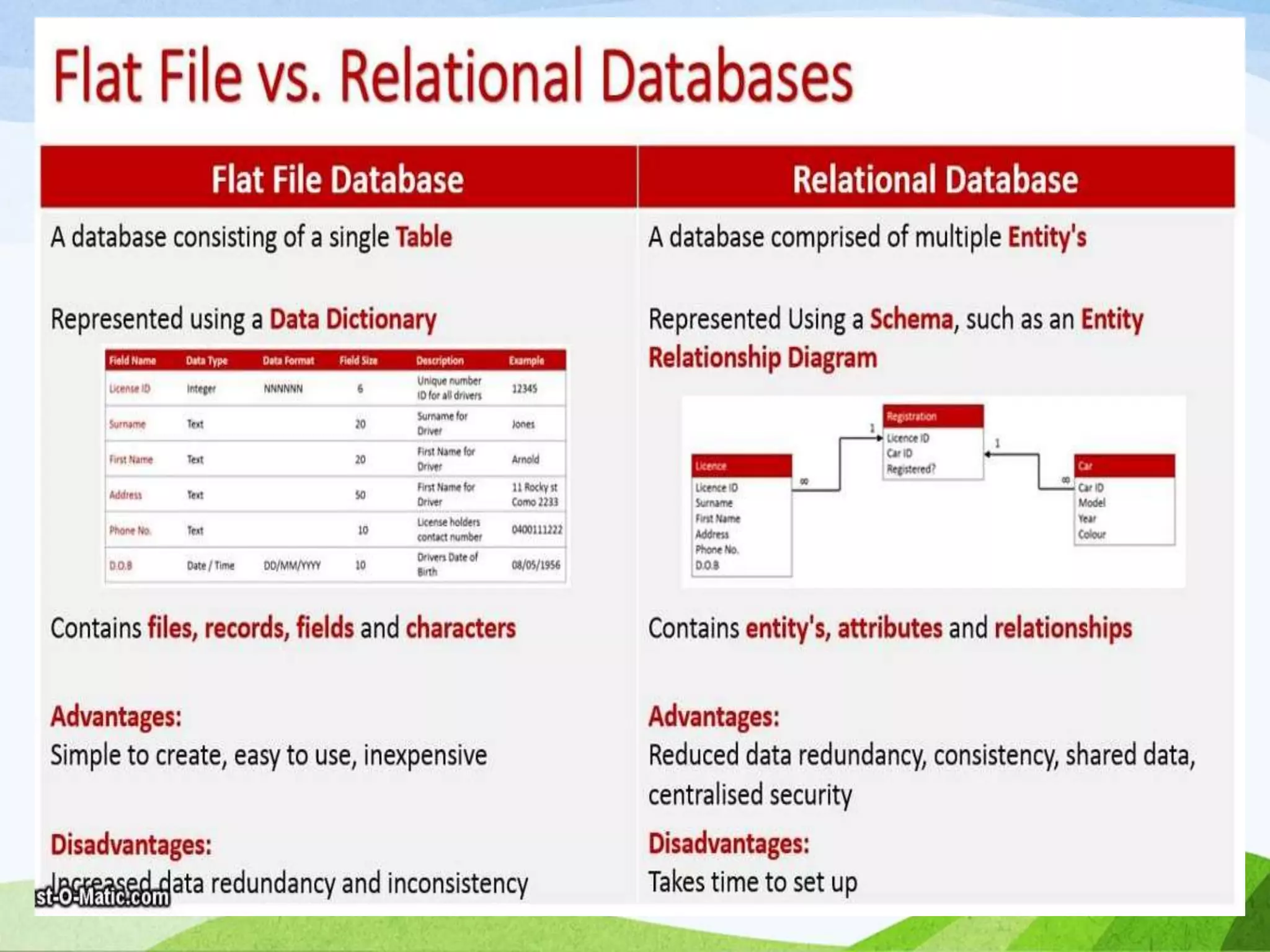
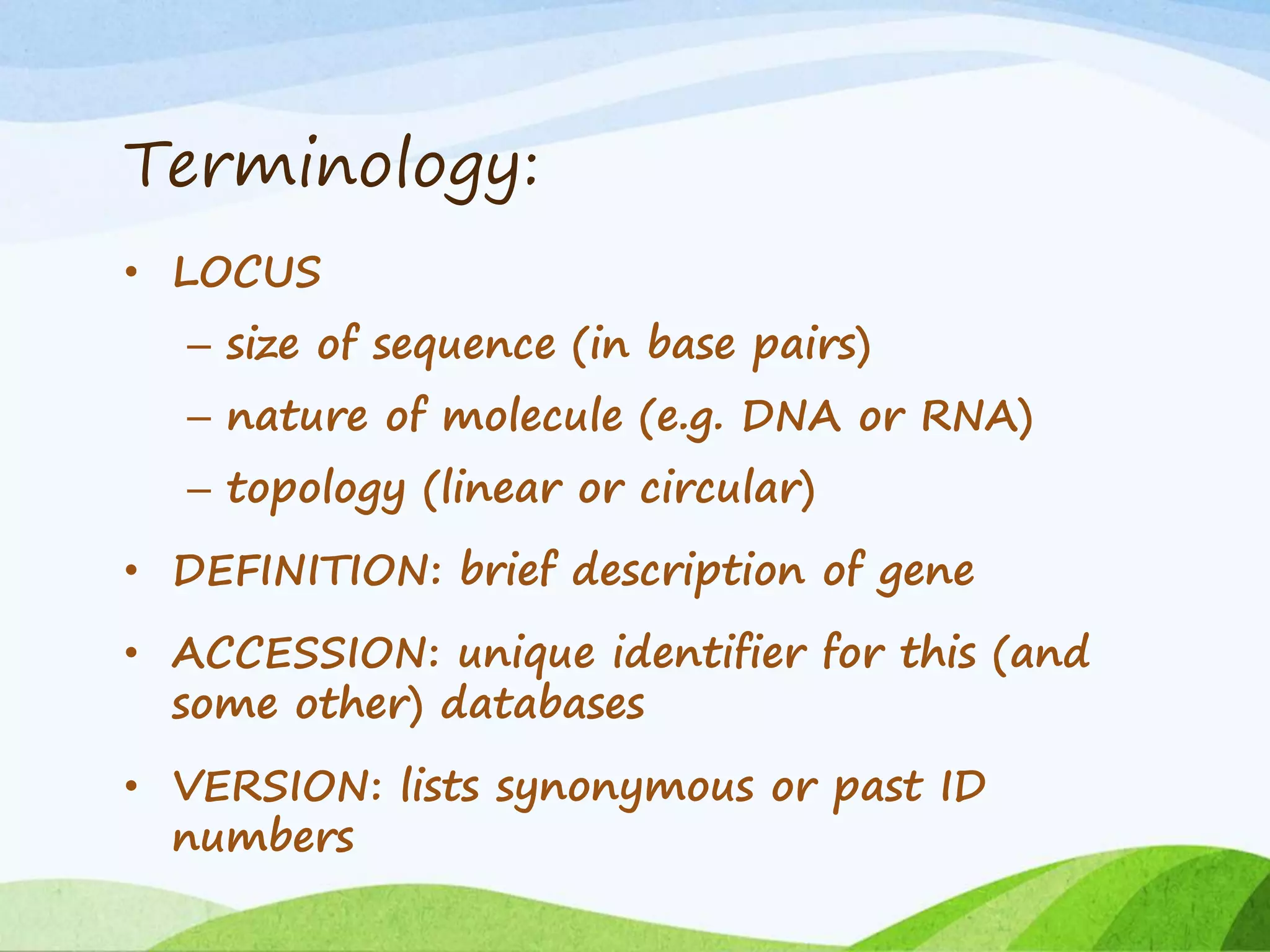
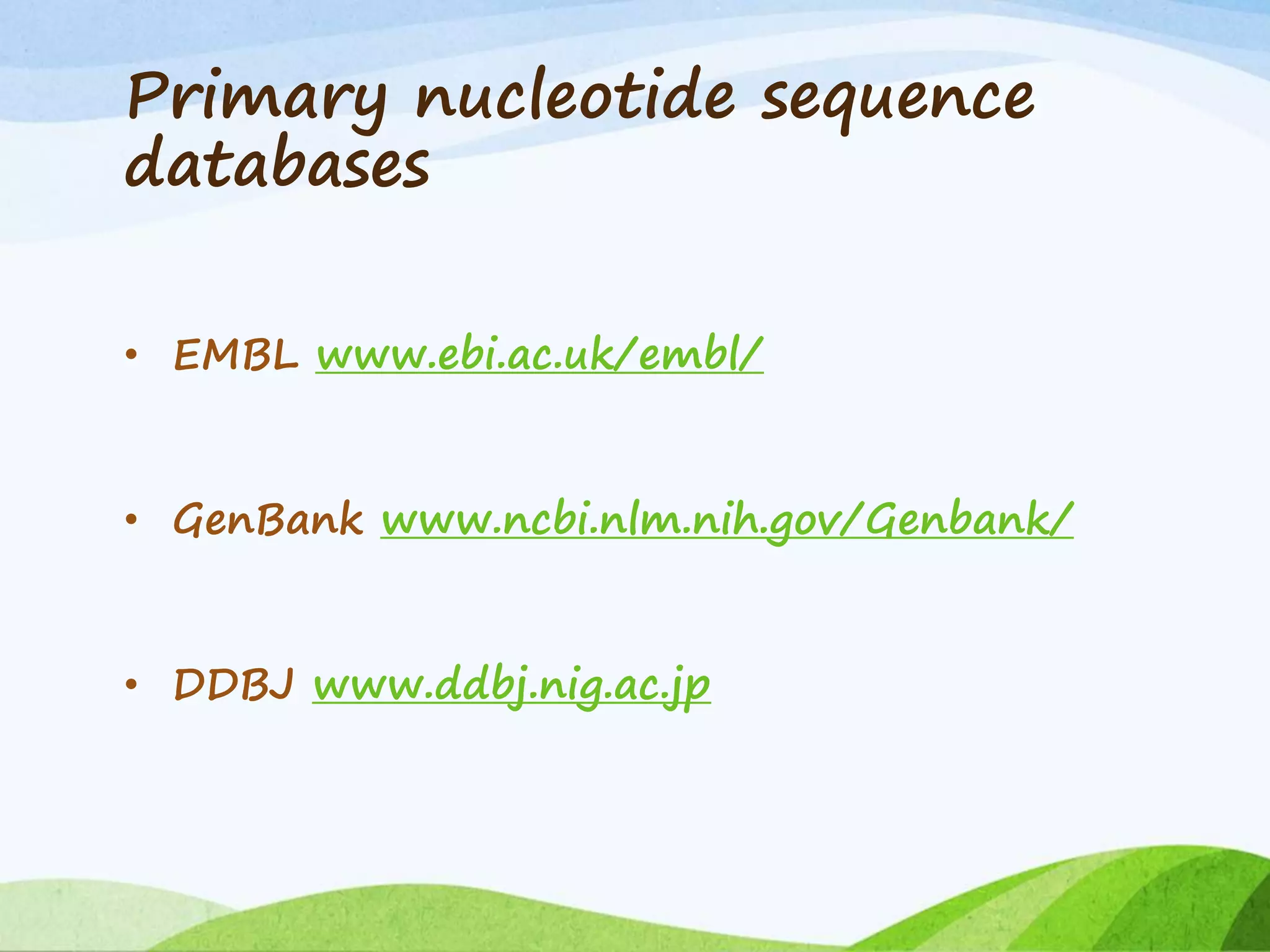
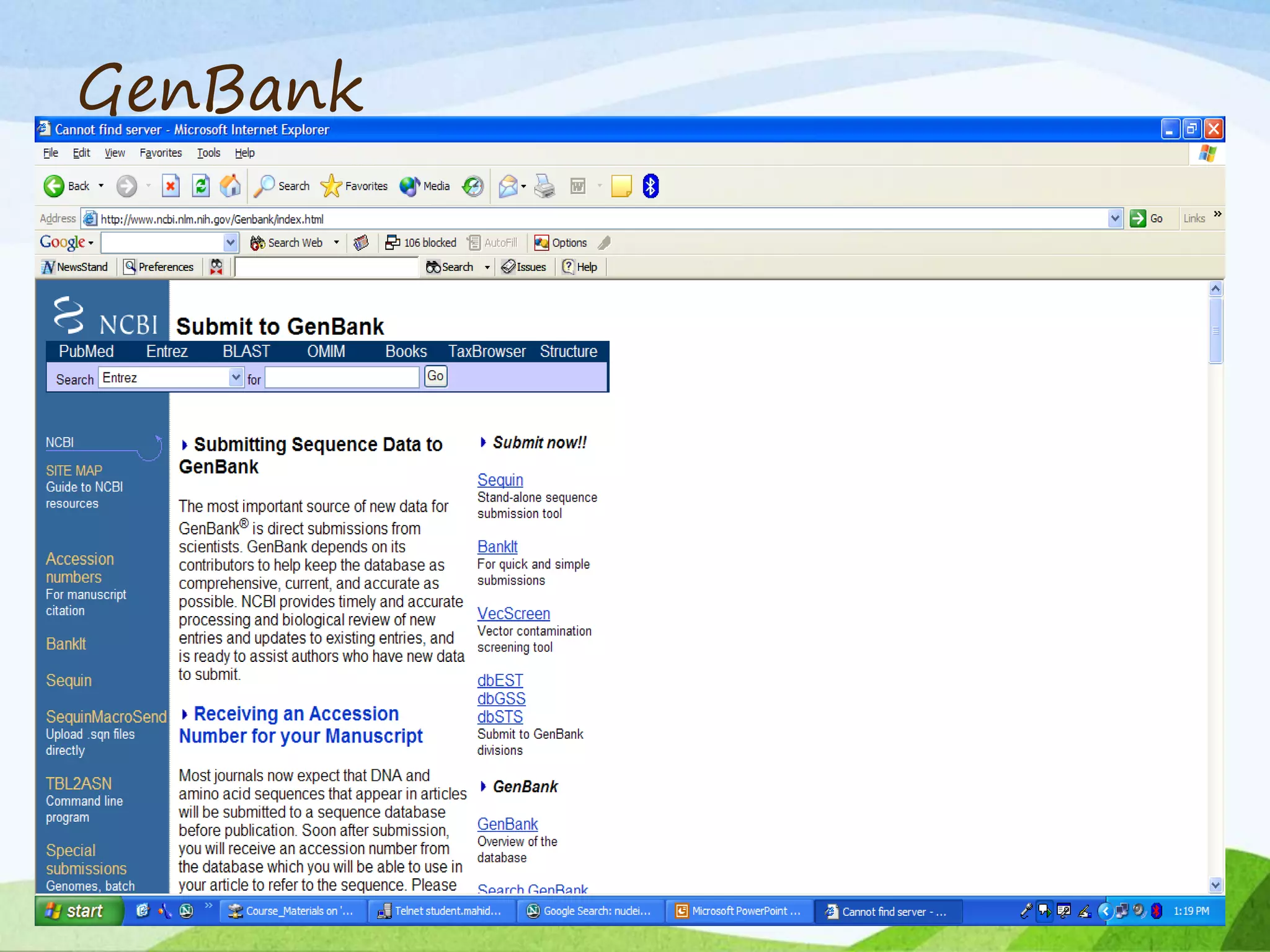
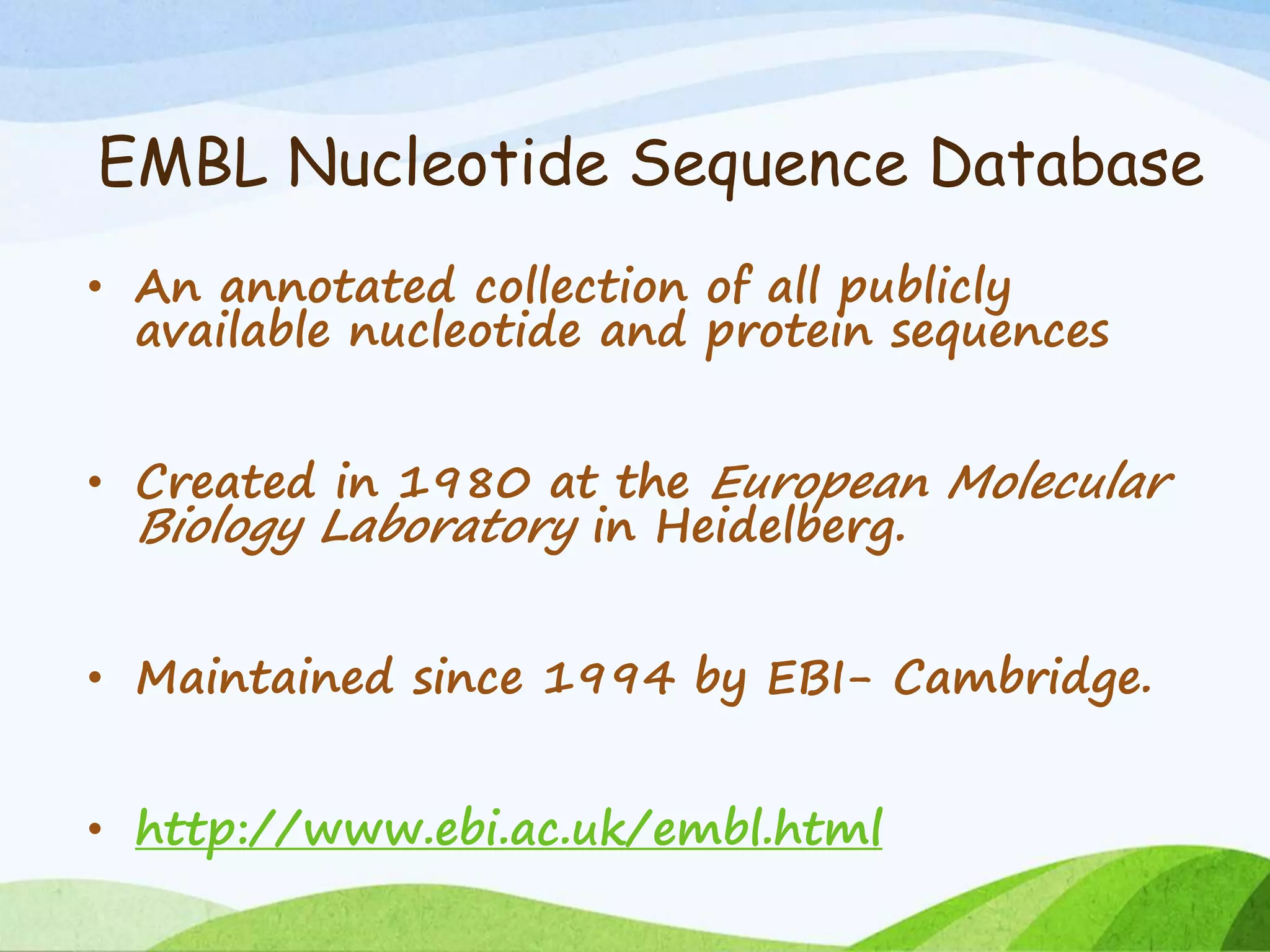
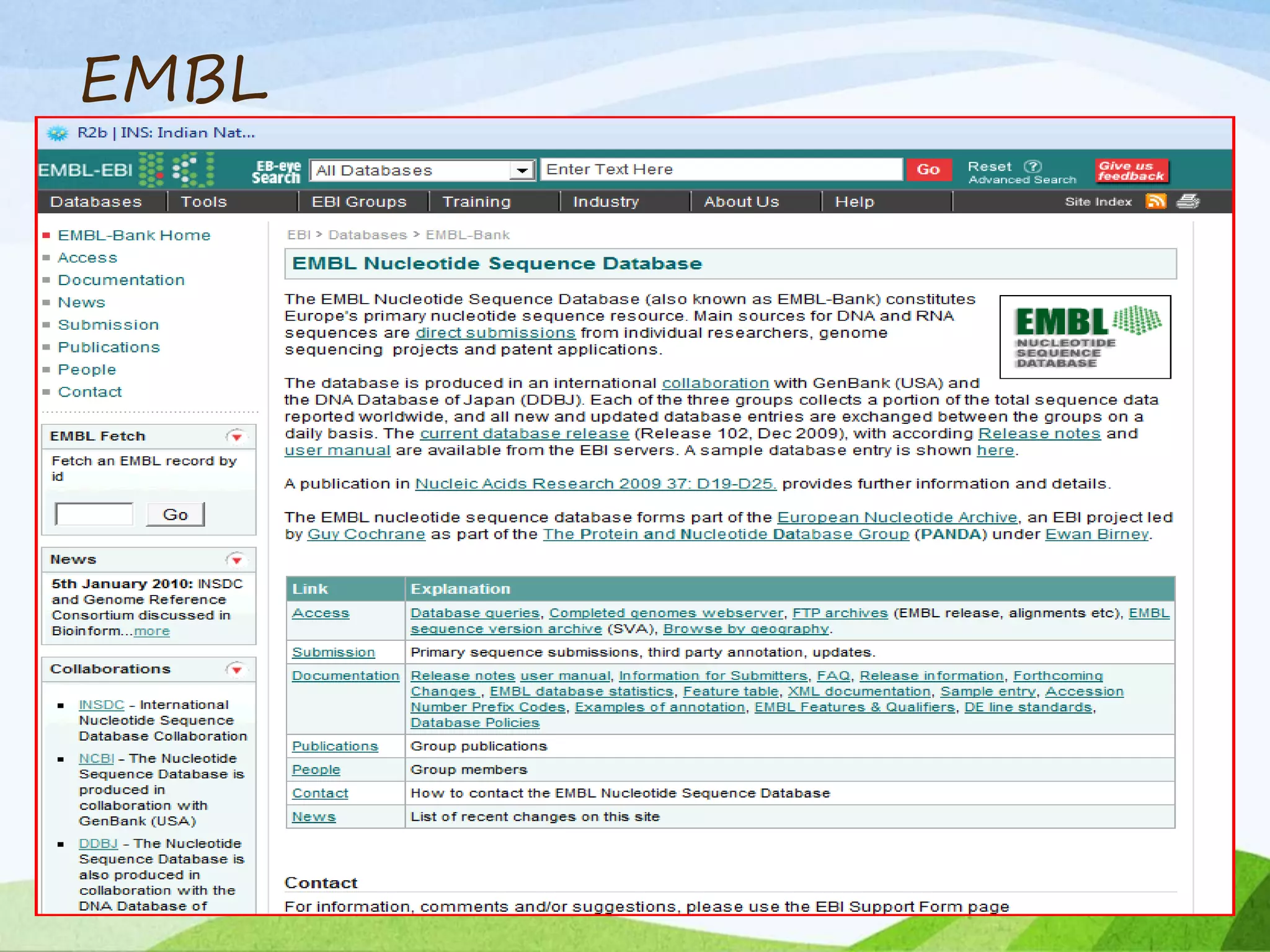
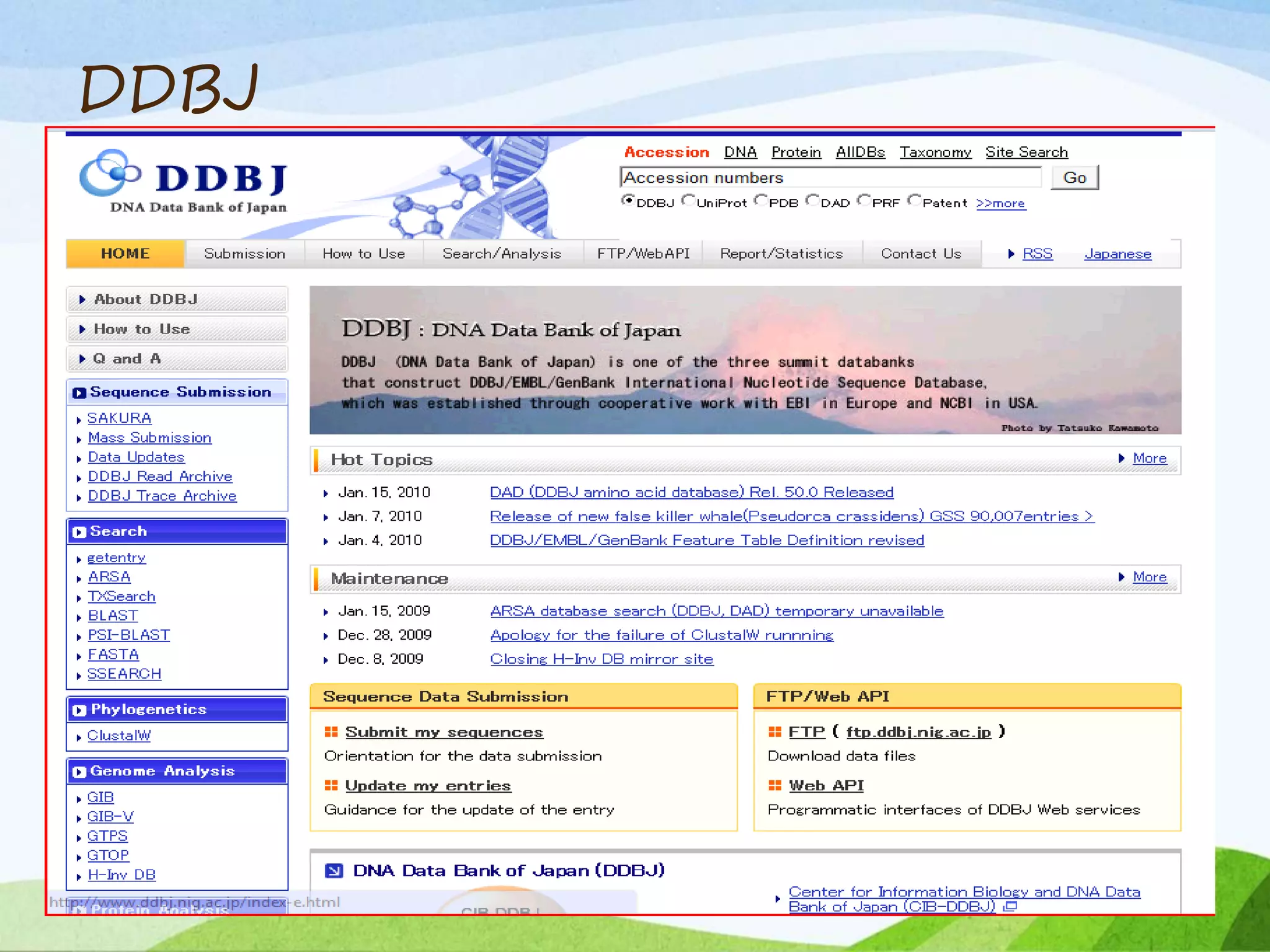
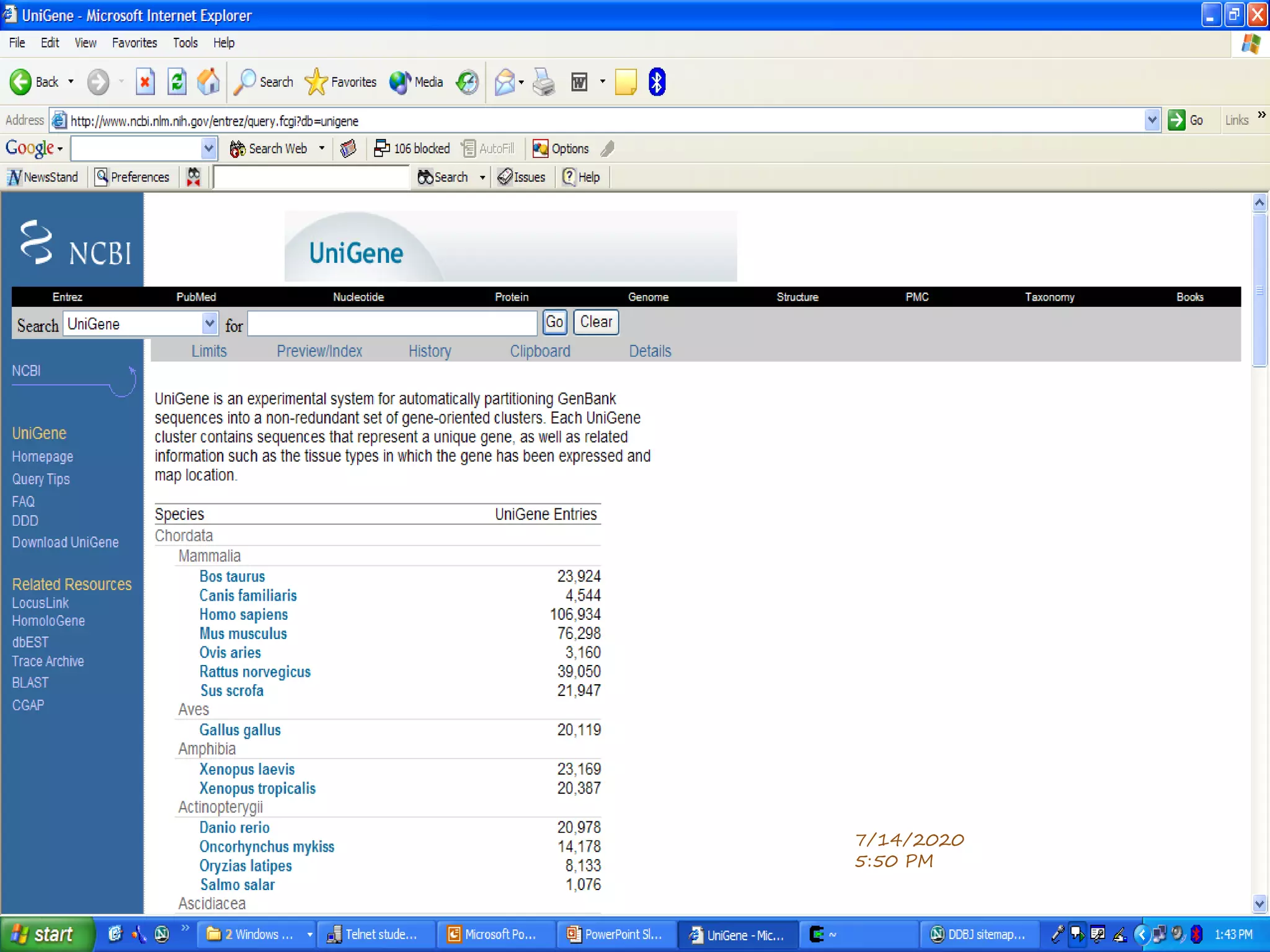
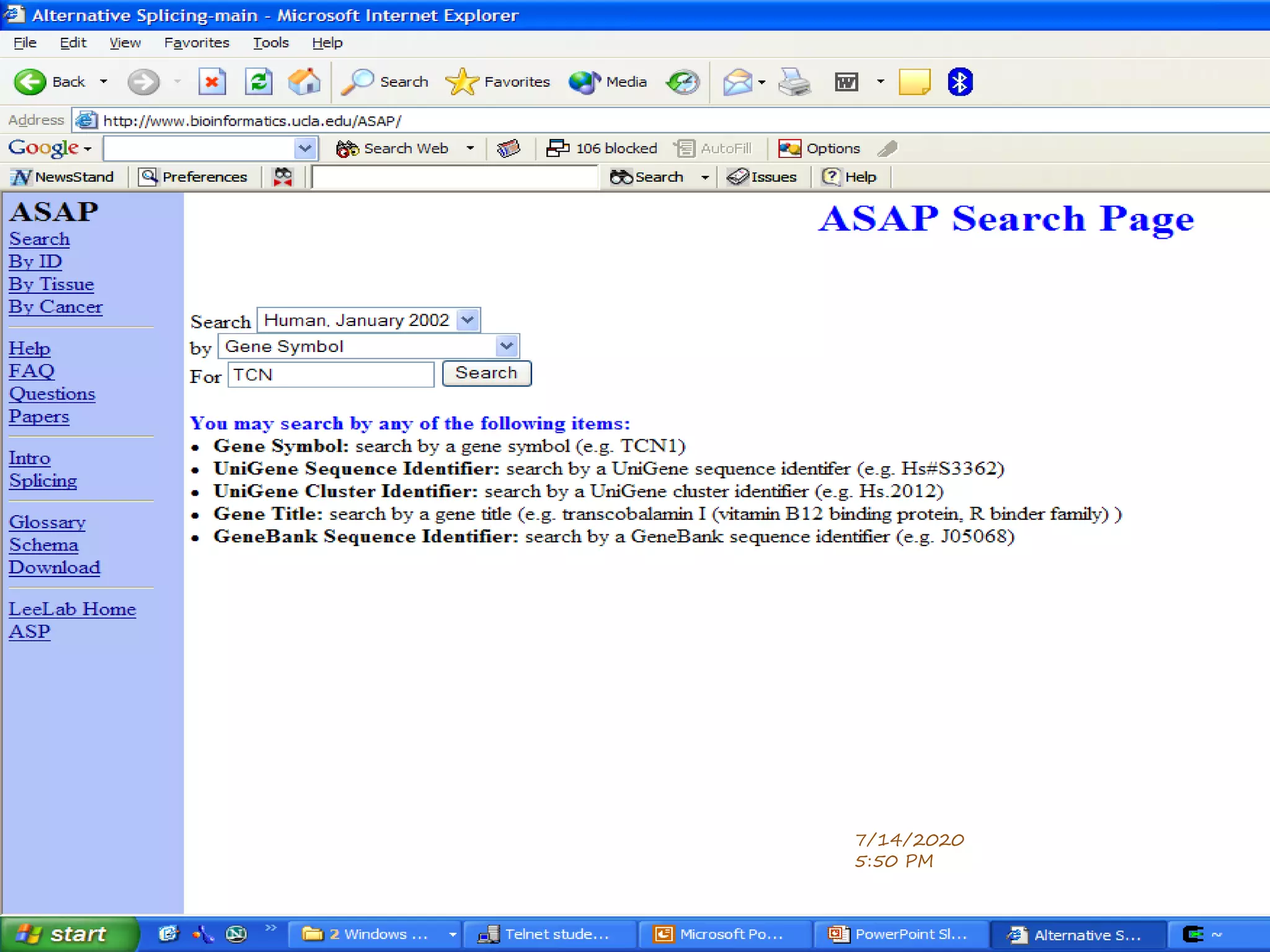
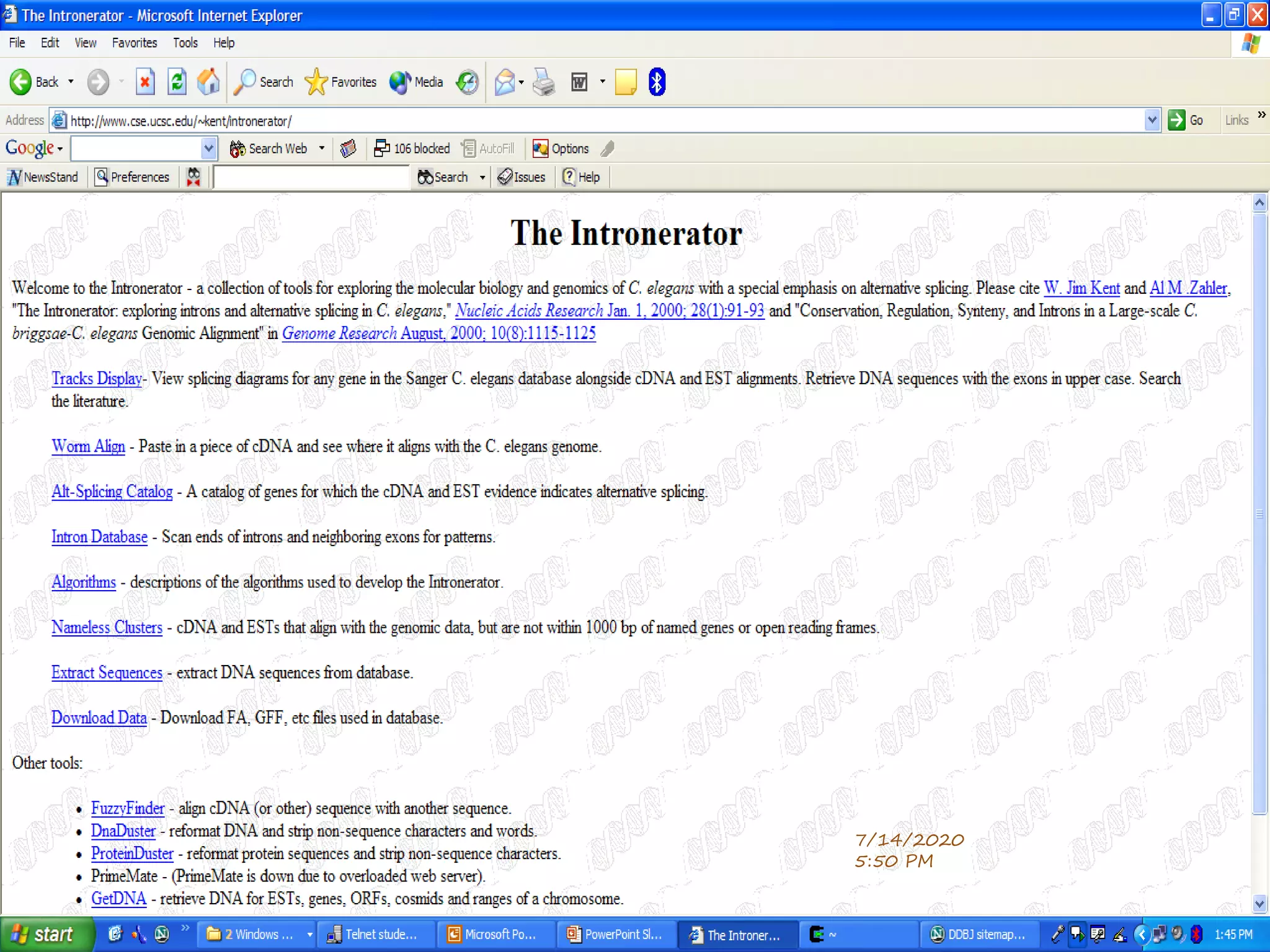
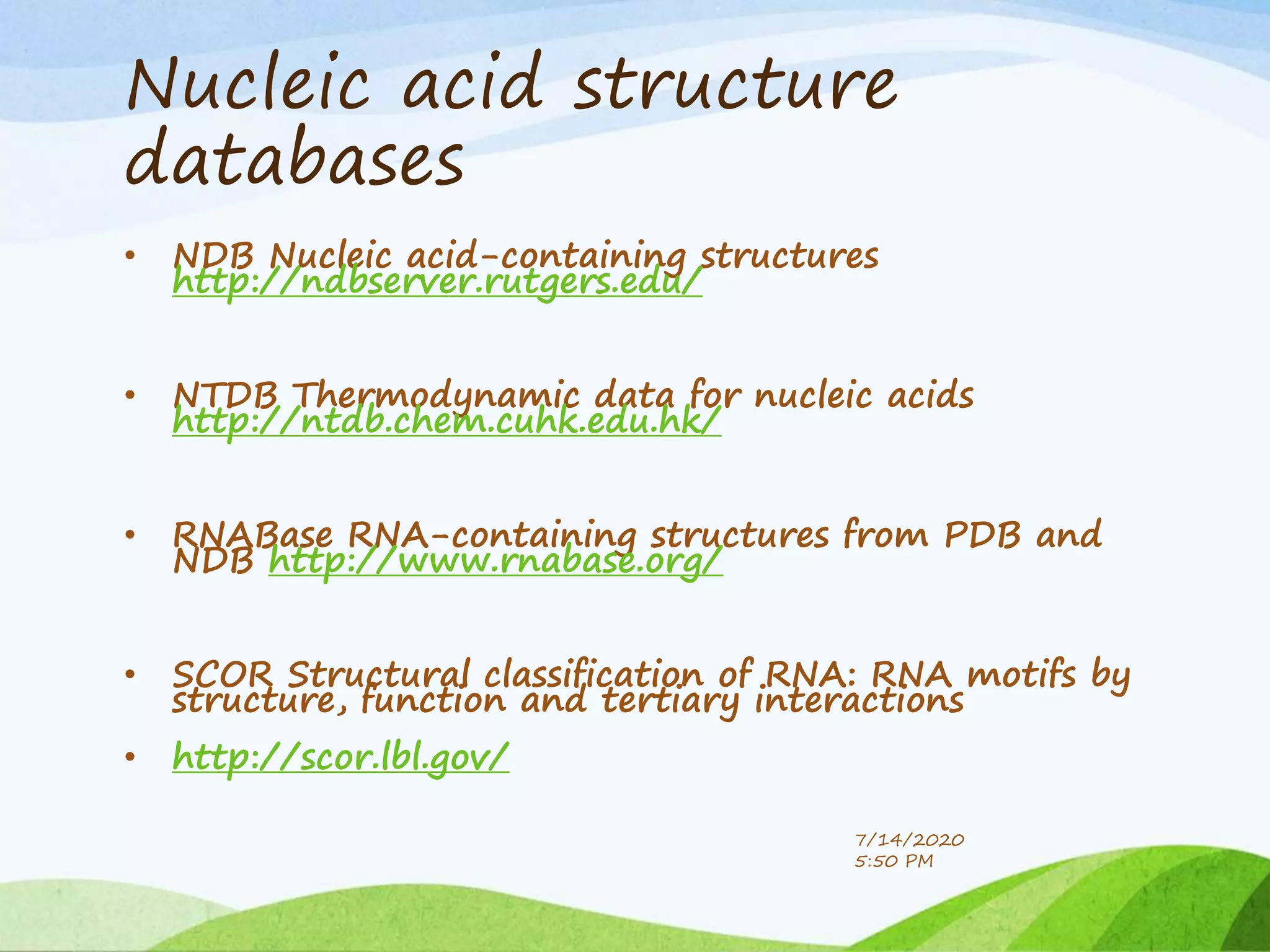
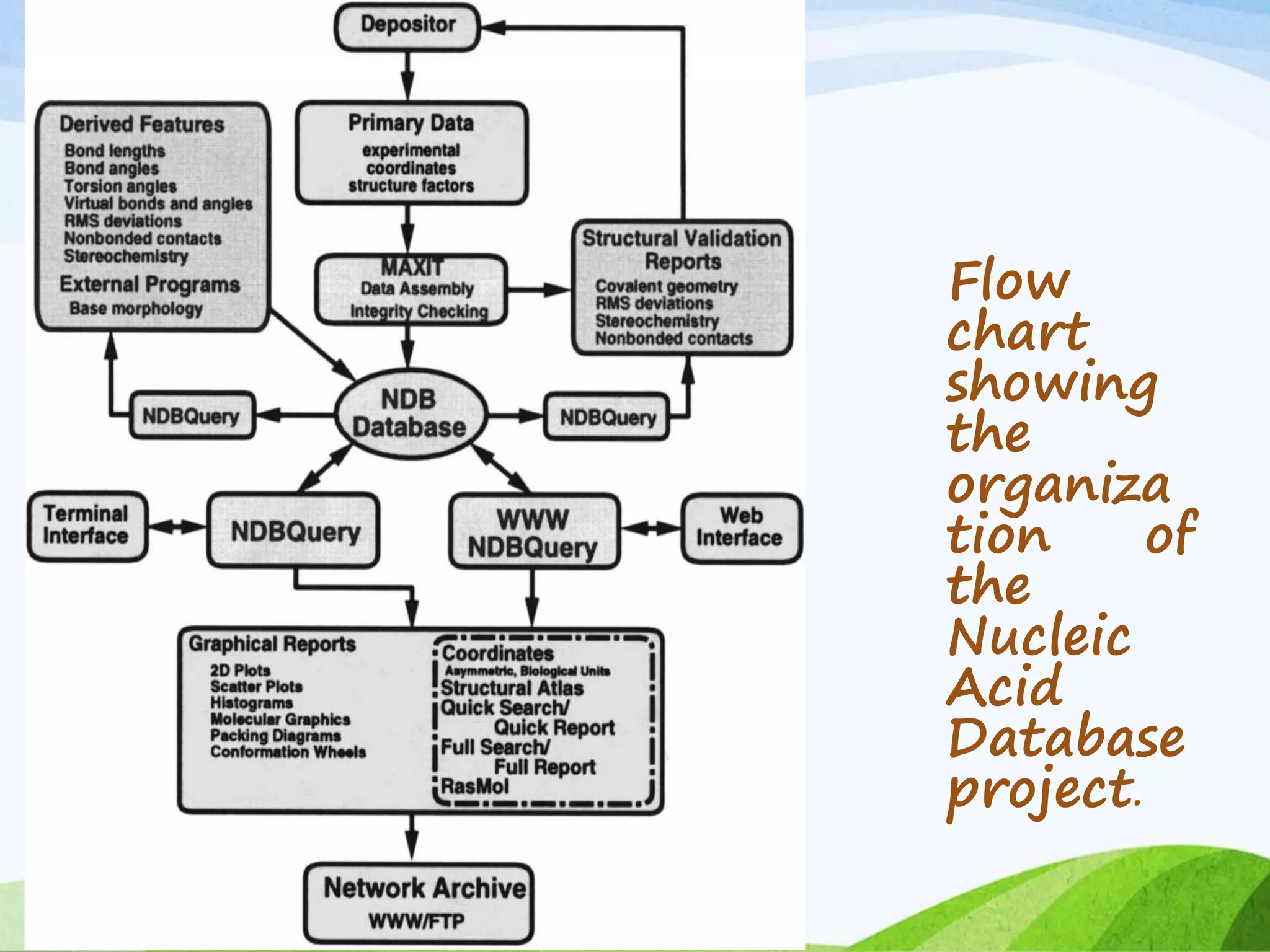
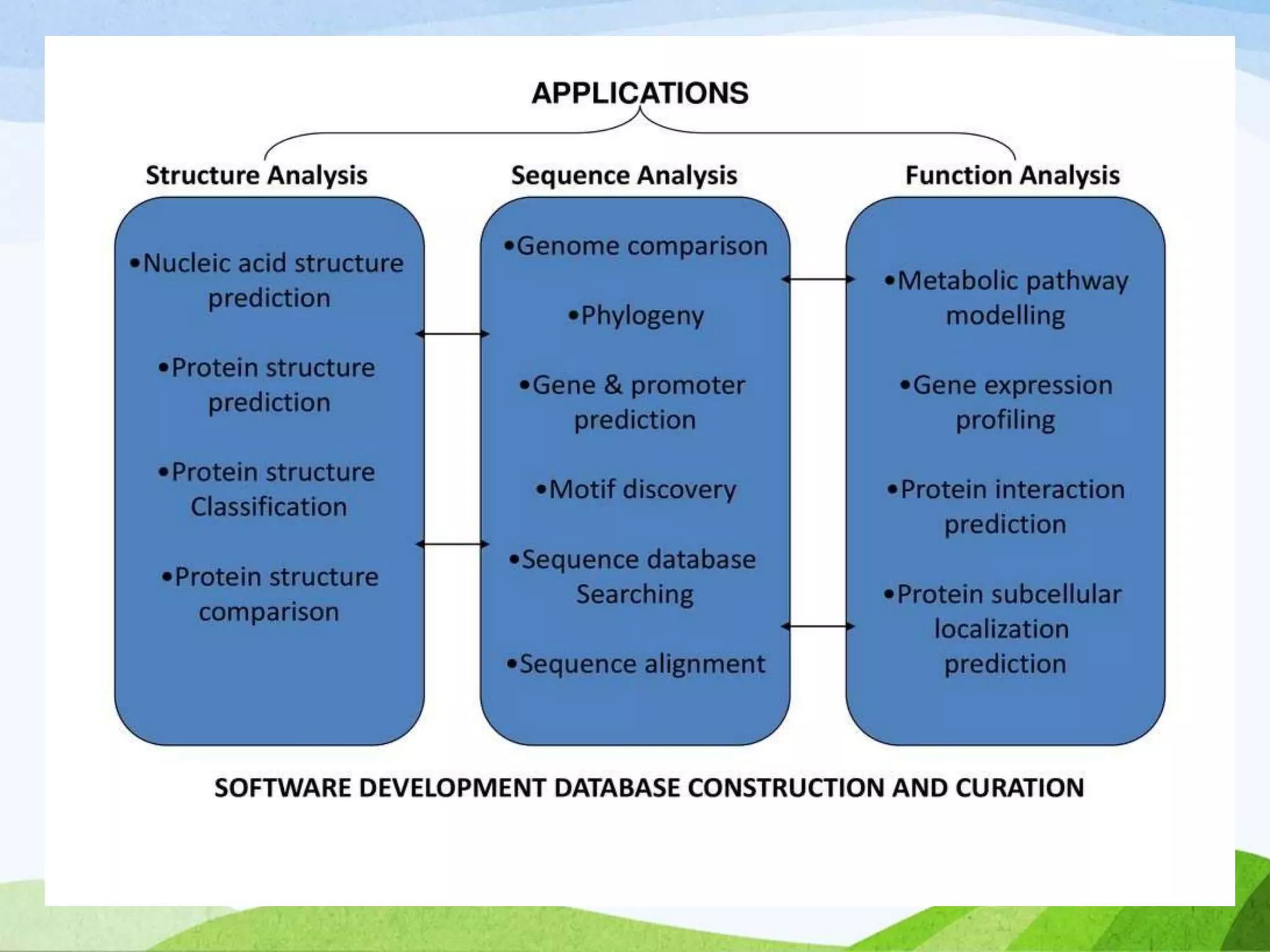
The document discusses biological databases that store and make available large datasets of biological data. It describes the aims of databases to store and communicate this data, make it available to scientists, and in a computer-readable format. The document classifies databases as primary, composite, or secondary. It provides details on the formats, availability, terminology, and examples of primary nucleotide sequence databases like GenBank, EMBL, and DDBJ. Derived databases and tools for sequence retrieval are also summarized.