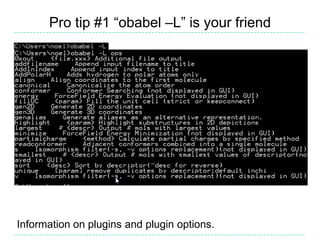
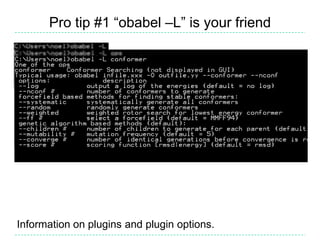
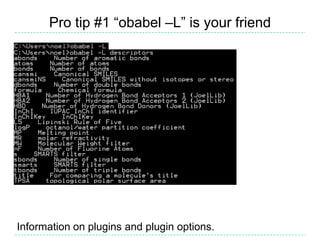
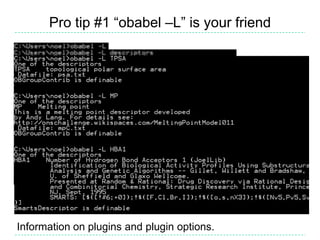
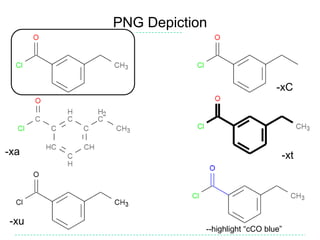
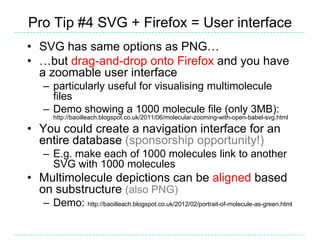
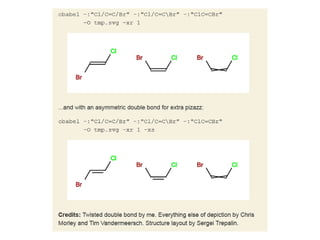
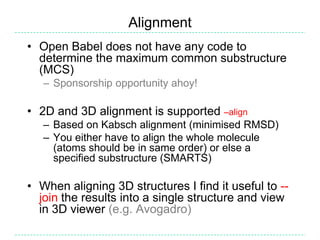
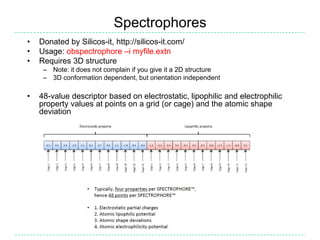
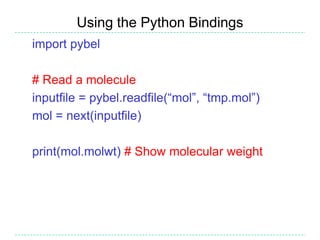
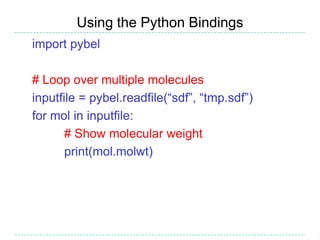
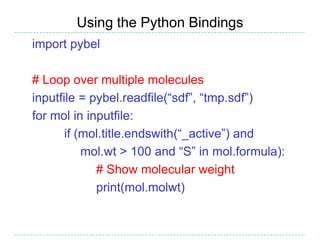
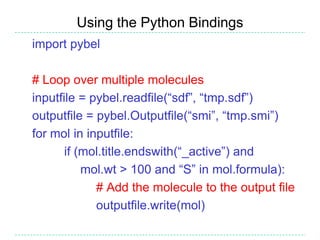
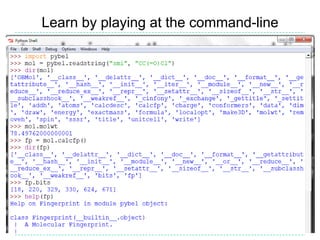
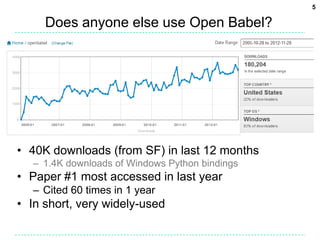
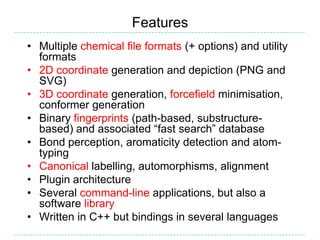
The document provides an overview of the Open Babel chemical toolbox. It discusses that Open Babel is an open source project for chemical file format conversion and manipulation. It has been in development since 2001 and has over 40,000 downloads per year. It supports multiple chemical file formats and has features for 2D/3D structure generation, fingerprints, reaction searching, and programming via its plugin architecture and language bindings.





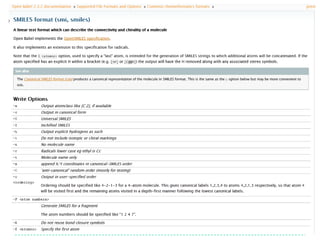

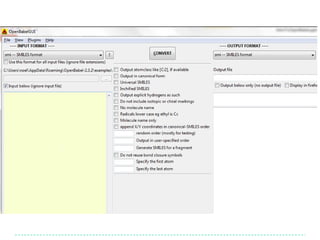
![SMILES output options
1. Add explicit Hs
Note that atom order is > obabel -:CC(=O)Cl –osmi 2. Show them in the
preserved CC(=O)Cl output
> obabel -:CC(=O)Cl –osmi –xh -h
Make atom 3 the first
[CH3]C(=O)Cl
atom… > obabel -:CC(=O)Cl -osmi -xf 3
O=C(C)Cl
…and atom 1 the > obabel -:CC(=O)Cl -osmi -xf 3 –xl 1
last O=C(Cl)C
> obabel -:CC(=O)Cl -:CC(=O)Cl -osmi -xC
ClC(=O)C Random order
O=C(Cl)C
> obabel -:CC(=O)Cl -osmi -xF "2 4"
CCl
Fragment SMILES for the fragment
composed of atoms 2 and 4
Take home message: Look through the list of options for file formats
which you frequently use (and request new options!)](https://image.slidesharecdn.com/introtoopenbabel-121204042028-phpapp01/85/Intro-to-Open-Babel-14-320.jpg)