Embed presentation
Download as PDF, PPTX

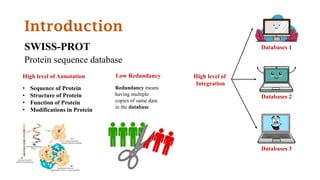
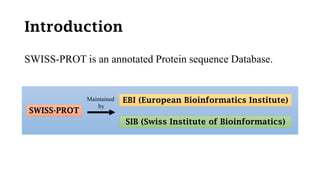
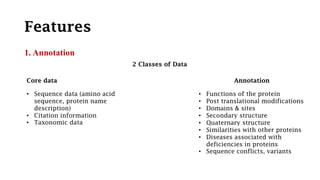
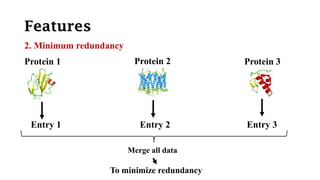
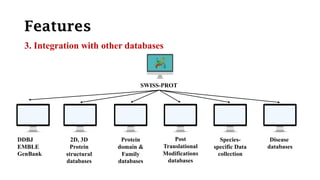
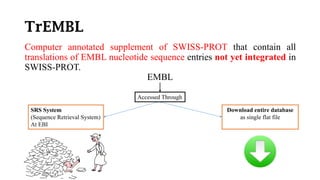
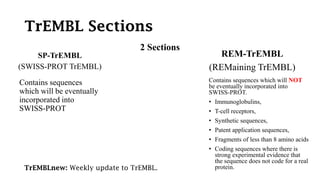
SWISS-PROT is a curated protein sequence database that provides a high level of annotation and low redundancy. It contains information on the sequence, structure, function, and modifications of proteins. The database is maintained by the European Bioinformatics Institute and Swiss Institute of Bioinformatics. It integrates with other databases containing structural, domain, post-translational, and disease information.







