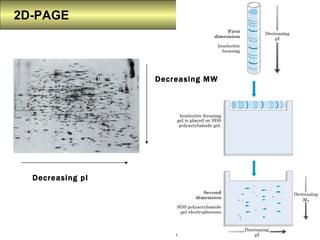
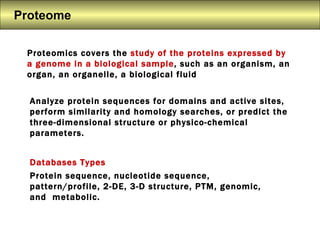
S.Prasanth Kumar is a bioinformatician who studies proteomics, 2D-PAGE, and proteome databases. Proteomics involves the study of proteins expressed by a genome through analysis of protein sequences, structures, modifications, and interactions. Major databases include Swiss-Prot, which contains annotated protein sequences, and TrEMBL, which contains automatically generated sequences. Other databases contain information on protein families and domains, nucleotide sequences, 2D-PAGE gel images, and post-translational modifications.
![S.Prasanth Kumar, Bioinformatician Proteomics 2D-PAGE & Proteome Databases S.Prasanth Kumar Dept. of Bioinformatics Applied Botany Centre (ABC) Gujarat University, Ahmedabad, INDIA www.facebook.com/Prasanth Sivakumar FOLLOW ME ON ACCESS MY RESOURCES IN SLIDESHARE prasanthperceptron CONTACT ME [email_address]](https://image.slidesharecdn.com/proteomedatabases-110324014145-phpapp01/75/Proteome-databases-1-2048.jpg)

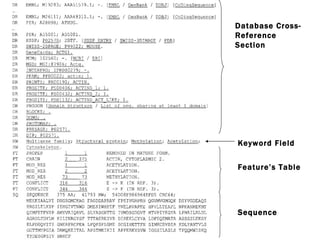
![Protein sequence databases SWISS-PROT an annotated universal sequence database, TrEMBL an automatically generated sequence database with repository character, which supplements SWISS-PROT. SWISS-PROT [http://www.expasy.org/sprot/] a curated protein sequence database which provides a high level of annotation Types of Annotations: Description of a protein's function, its domain structure, PTMs, conflicts between literature references and variants. It also provides a minimal level of redundancy , a high level of integration with other bio molecular databases , and an extensive external documentation. (Created in 1986:Main Host-ExPaSy)](https://image.slidesharecdn.com/proteomedatabases-110324014145-phpapp01/85/Proteome-databases-5-320.jpg)