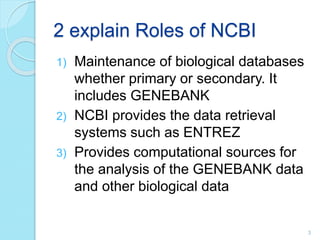
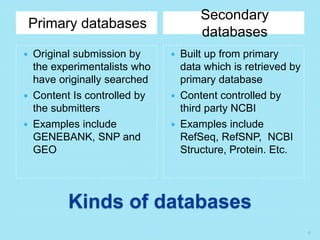
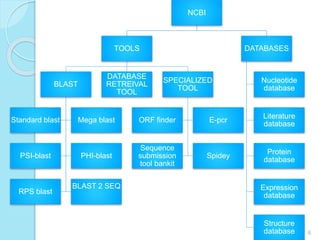
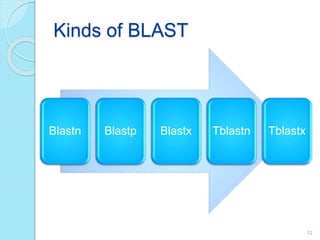
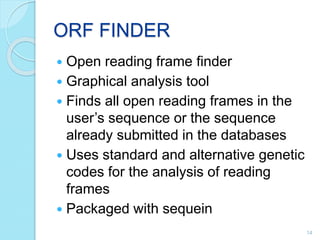
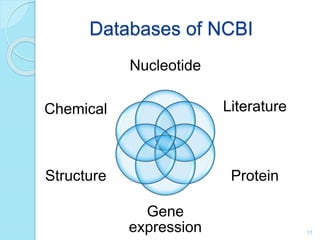
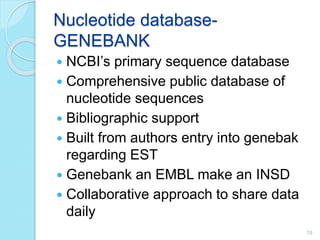
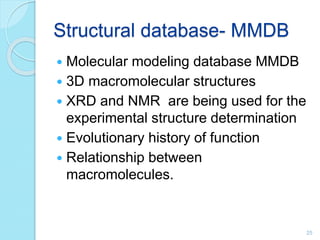
The document discusses the National Center for Biotechnology Information (NCBI), which maintains biological databases and provides bioinformatics tools. NCBI houses both primary databases directly submitted by researchers and secondary databases compiled from primary sources. Major databases include GenBank (nucleotide sequences), PubMed Central (biomedical literature), and reference sequence databases. Tools like BLAST, Entrez, and ORFfinder allow users to search and analyze sequence data. NCBI aims to make biomedical research data freely accessible worldwide.