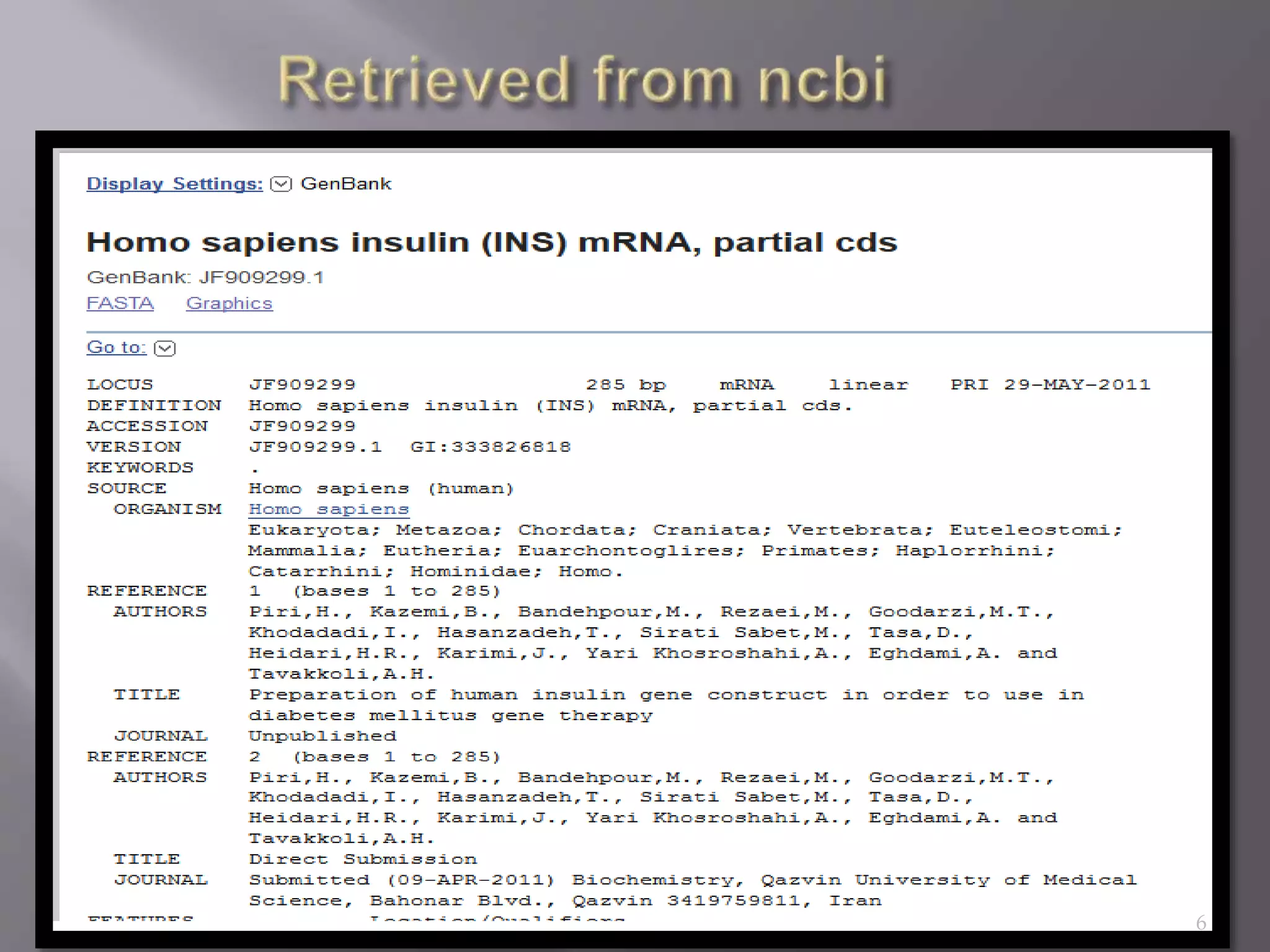
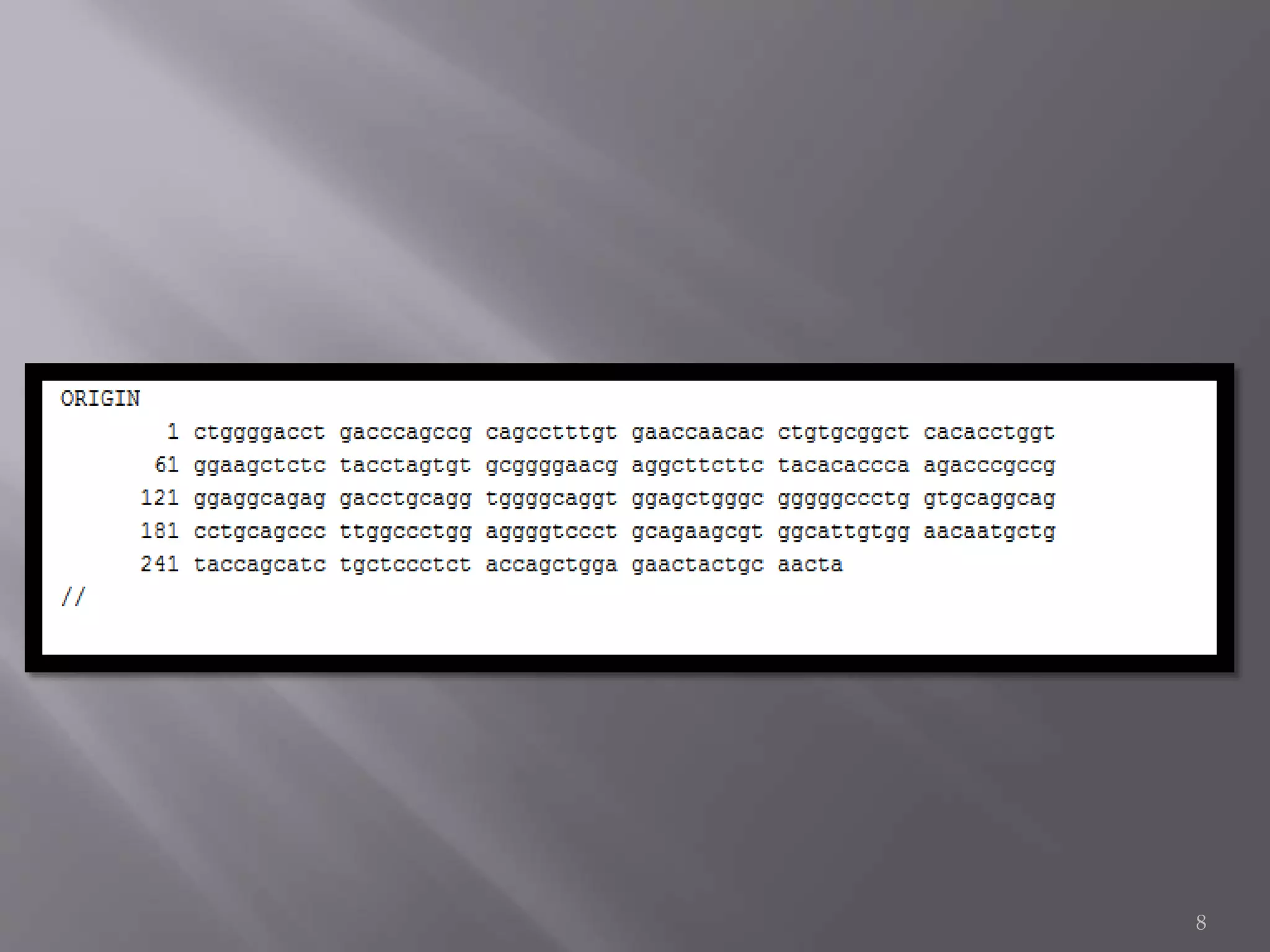
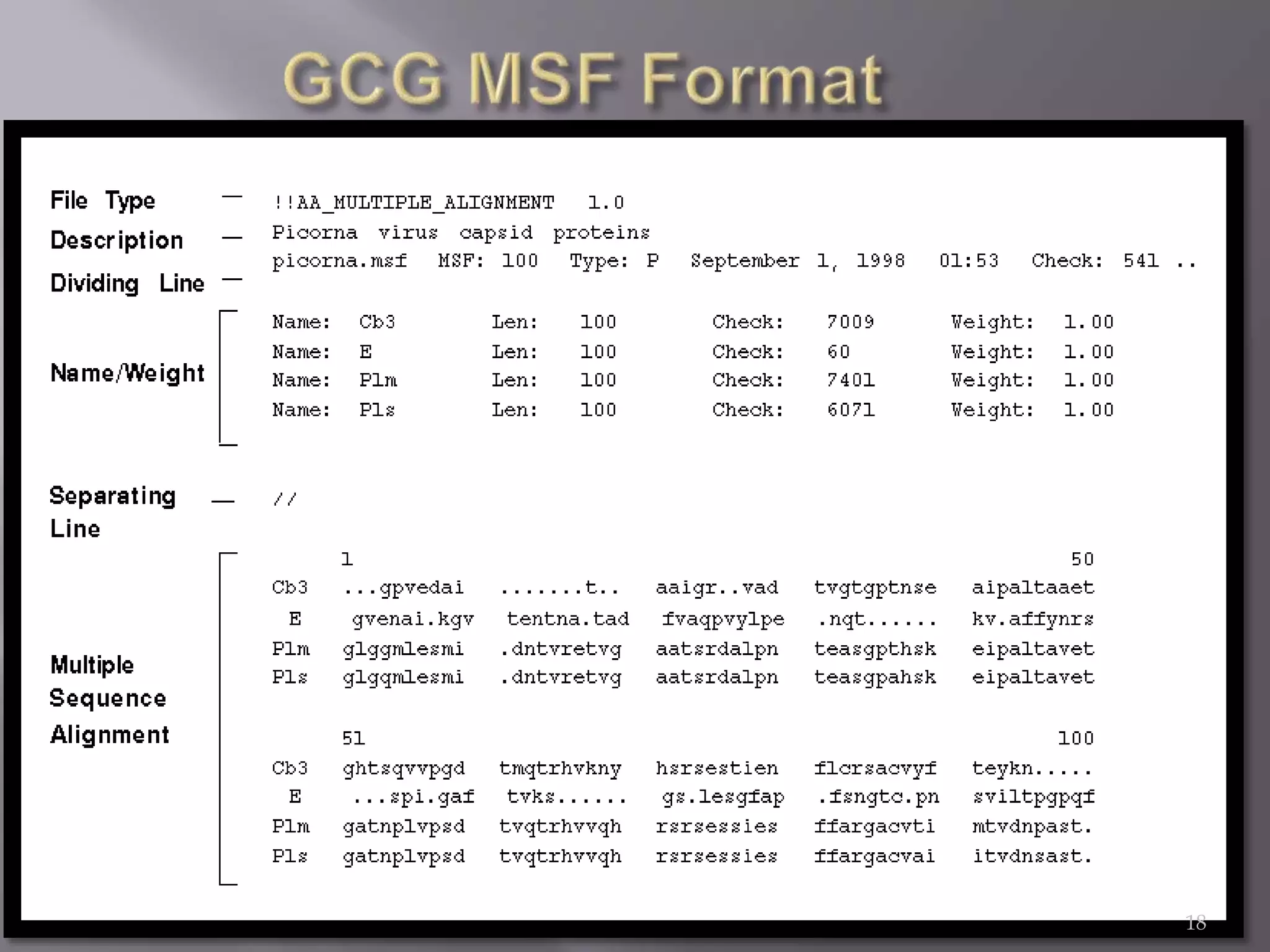
This document discusses different biological database file formats for storing sequence and molecular data. It describes several common formats including FASTA, Multi-FASTA, GeneBank flat-file format, GCG, EMBL, Clustal, and SWISS PROT. It provides details on the header, feature, and sequence sections of each format and how they represent genetic and protein information.