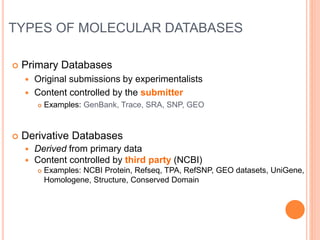
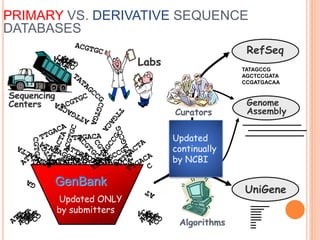
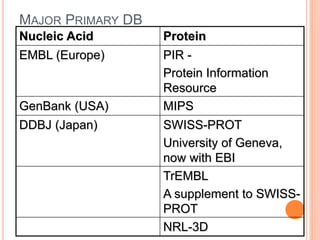
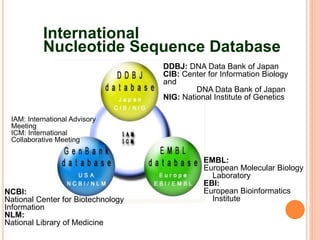
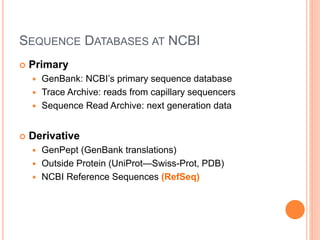
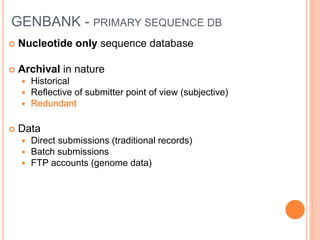
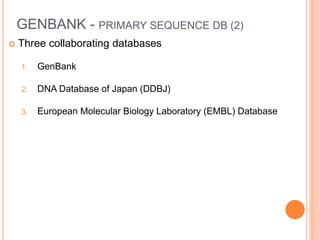
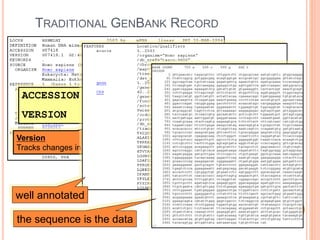
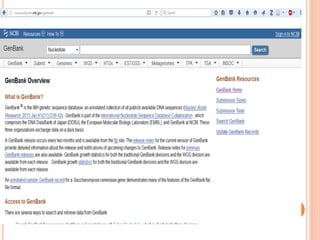
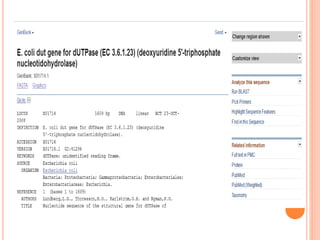
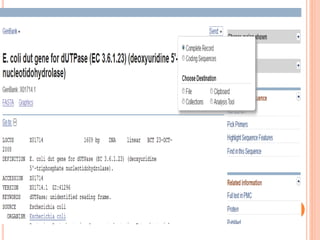
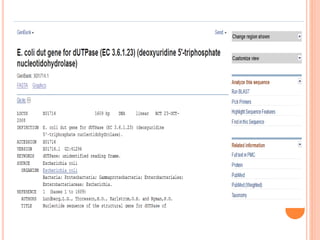
The document provides an overview of biological databases, their structure, and types, including primary and derivative databases. It highlights key features of data organization, the functionality of well-maintained databases, and the role of prominent databases like GenBank and Swiss-Prot. Additionally, it discusses the importance of data integration for revealing inter-related biological information.