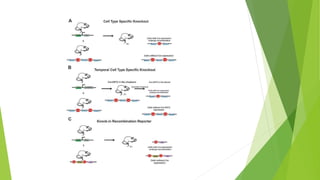
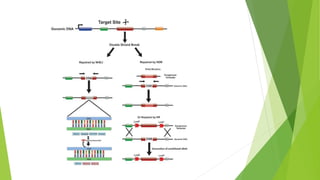
The document discusses gene targeting, a method to modify specific genes using a targeting vector and homologous recombination, enabling gene knockout or introduction for research and therapeutic purposes. It also details expressed sequence tags (ESTs), which are short sequences used in gene discovery and mapping, with implications for understanding gene expression and function. The significance of the work in gene targeting is highlighted by the 2007 Nobel Prize awarded to key researchers in this field.