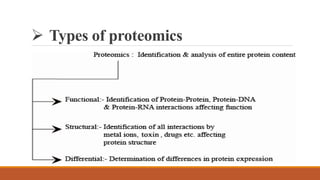
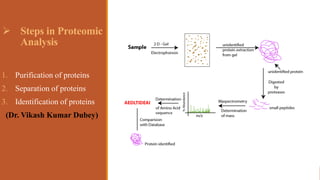
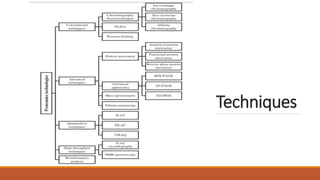
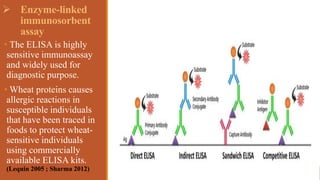
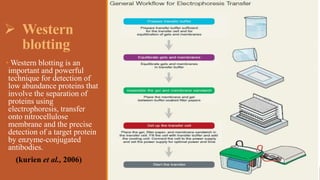
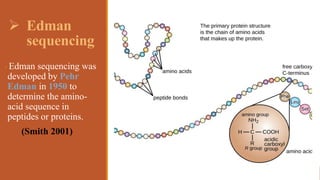
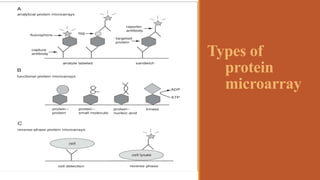
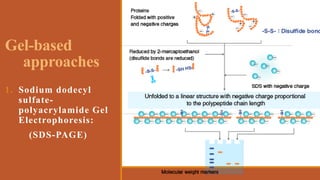
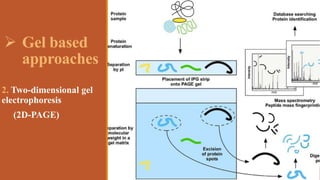
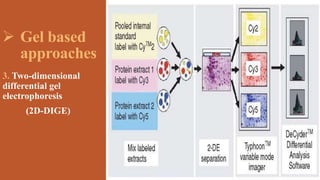
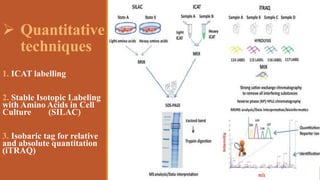
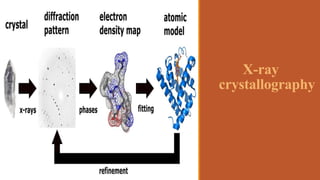
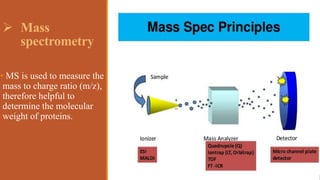
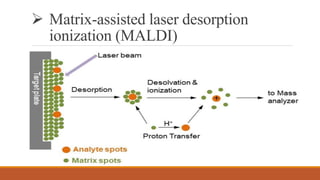
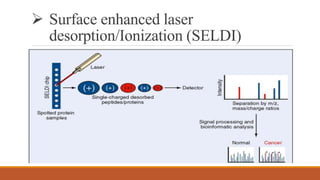
This document provides an overview of the field of proteomics. It lists the names and student IDs of 10 students in Group #3 who will study proteomics. It then outlines the topics to be covered, including an introduction defining proteomics and the proteome, a brief history of the field, techniques used in proteomics like mass spectrometry and gel electrophoresis, advantages and disadvantages, and applications in fields like oncology and agriculture.