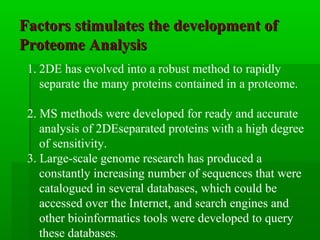
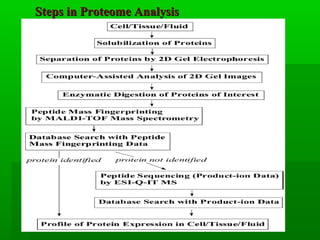
Proteome analysis studies the array of proteins expressed in a biological system under particular conditions. It can help understand cellular pathways and biological processes by characterizing protein complexes. Proteome analysis is also used to discover disease biomarkers for diagnostics and drug development by identifying protein expression changes. Key developments driving the field include improvements to 2D electrophoresis for separating proteins, mass spectrometry for analyzing separated proteins, and bioinformatics tools for searching protein databases. Common steps in proteome analysis involve separating proteins, digesting them into peptides, and identifying peptides and proteins using mass spectrometry techniques like MALDI-TOF-MS and ESI-Q-IT-MS.