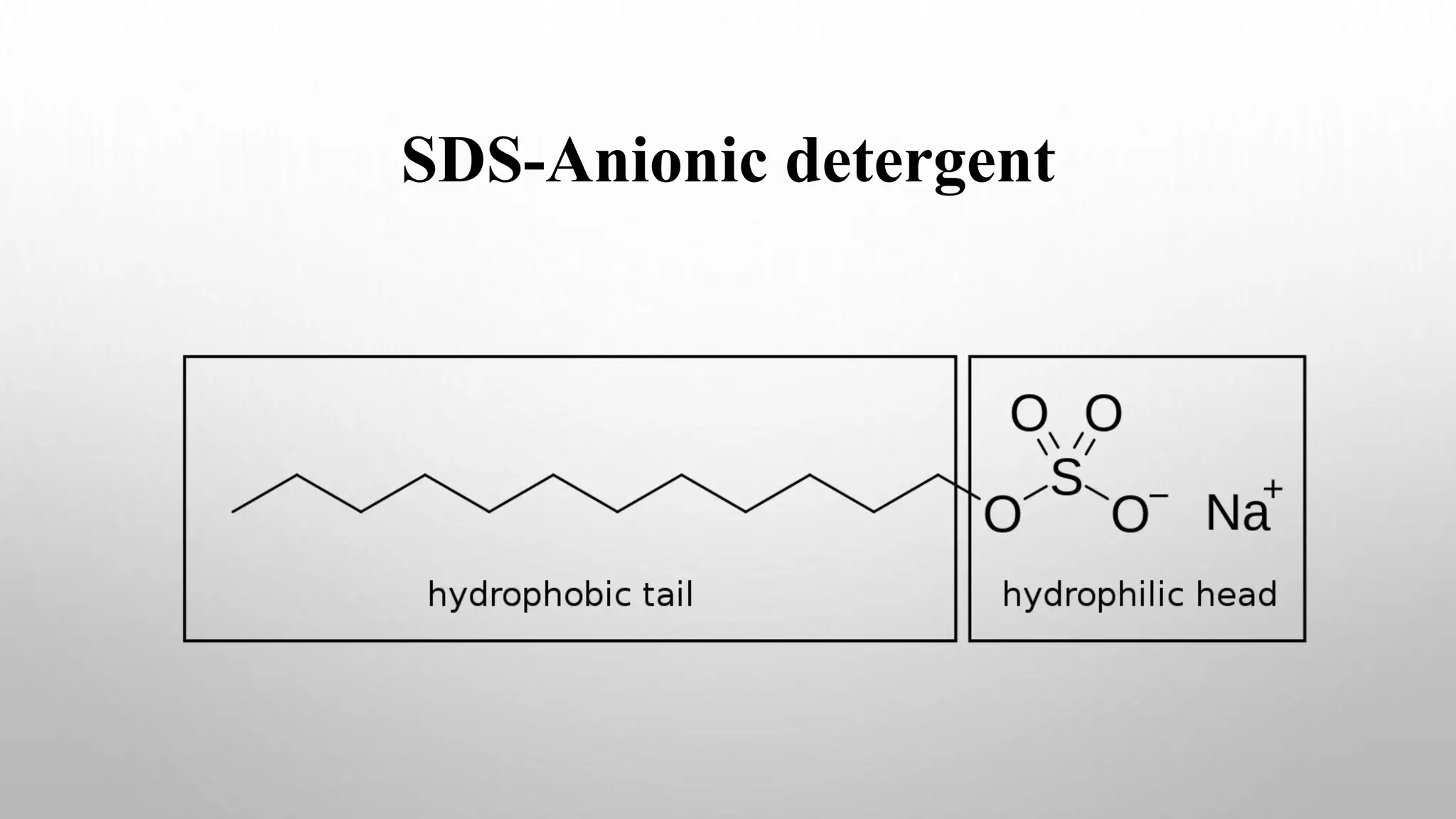
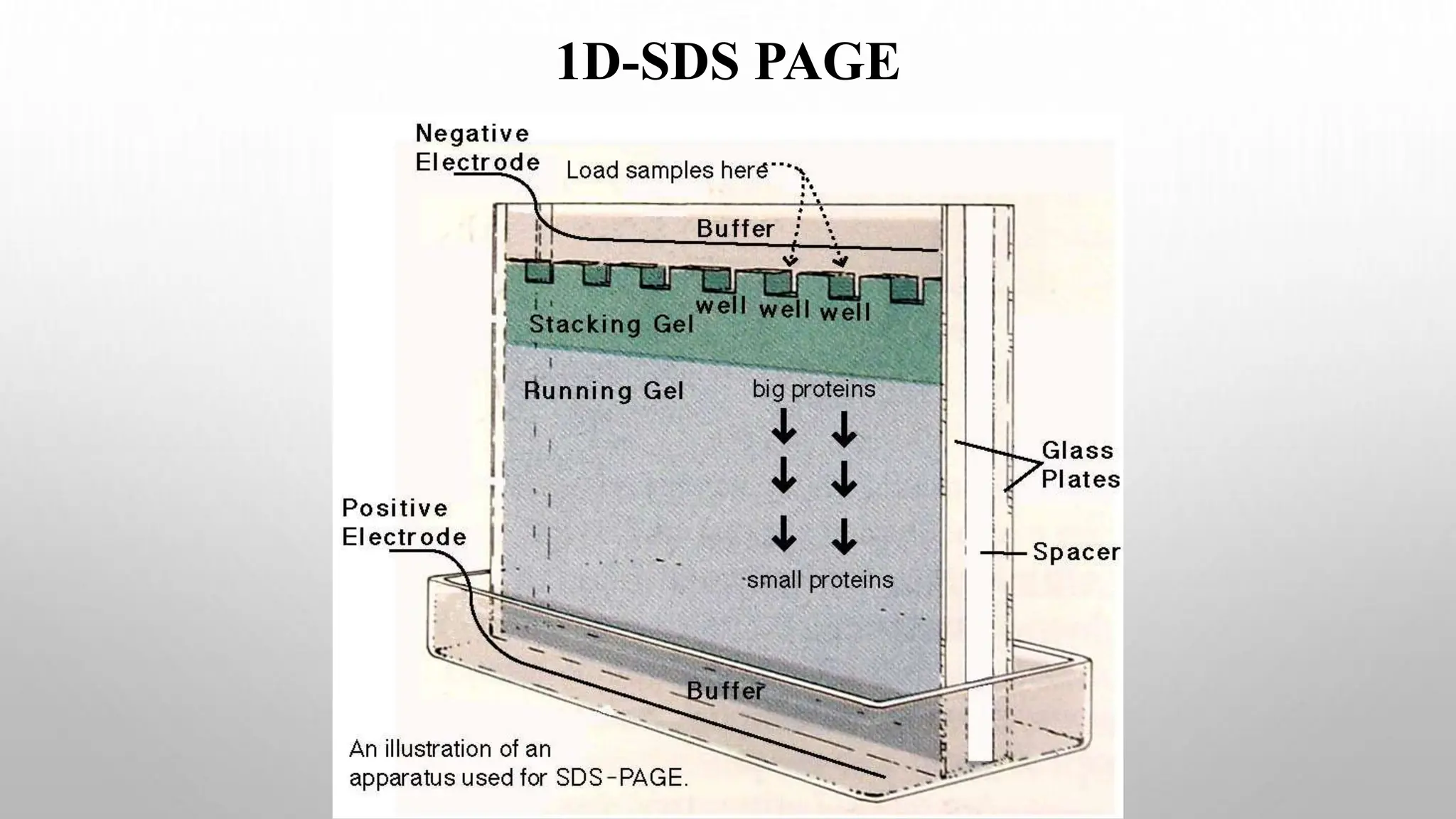
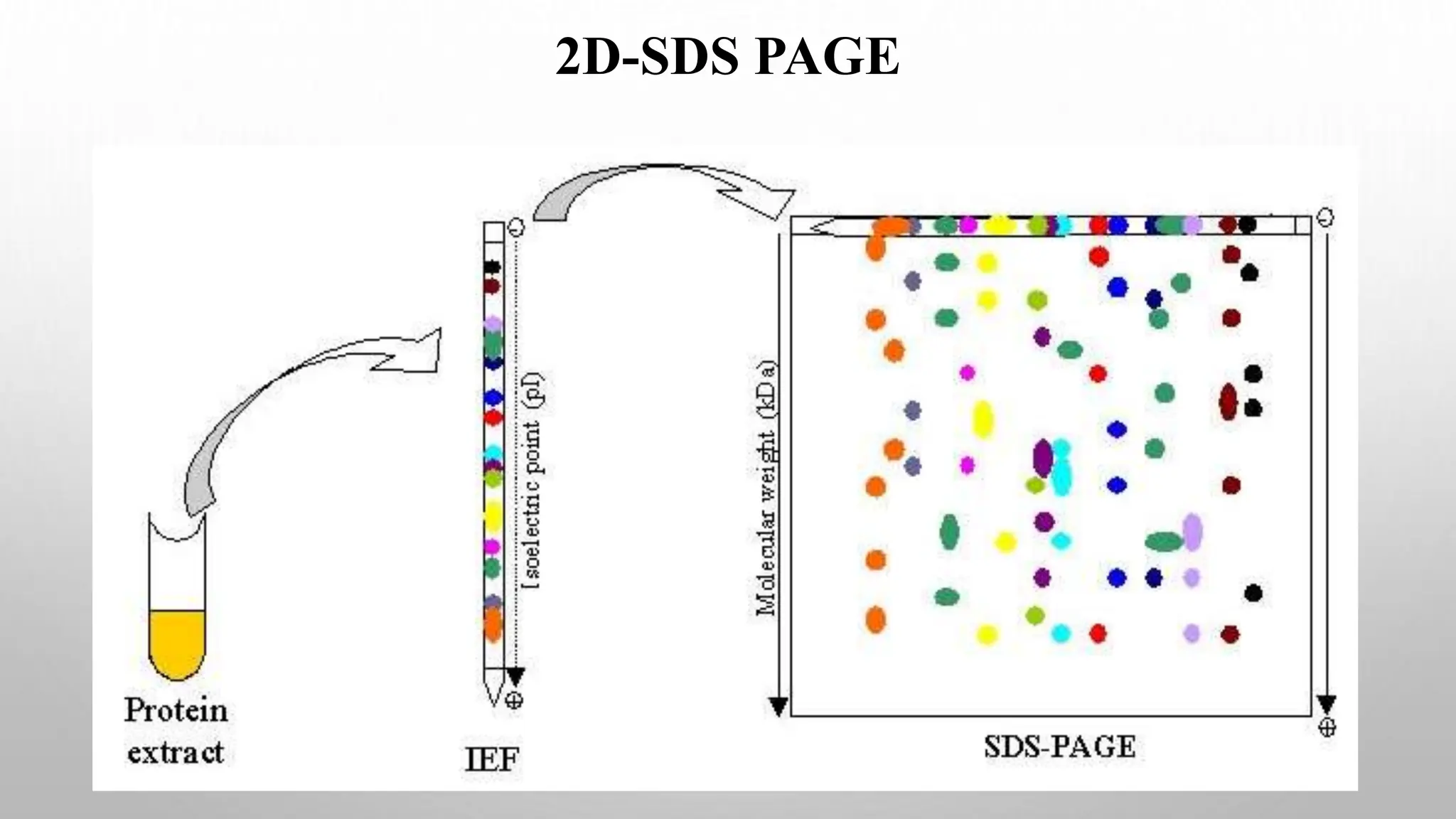
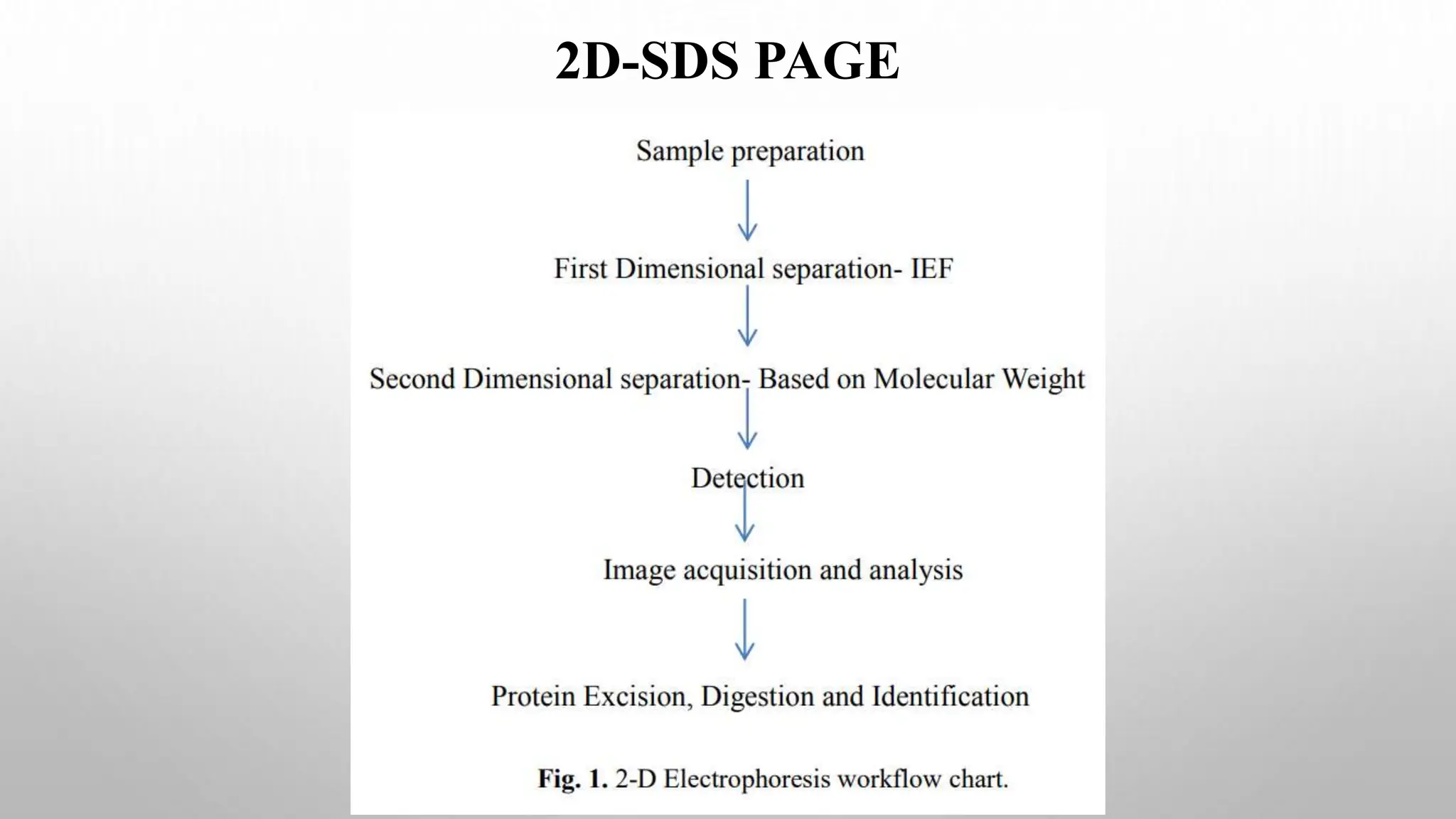
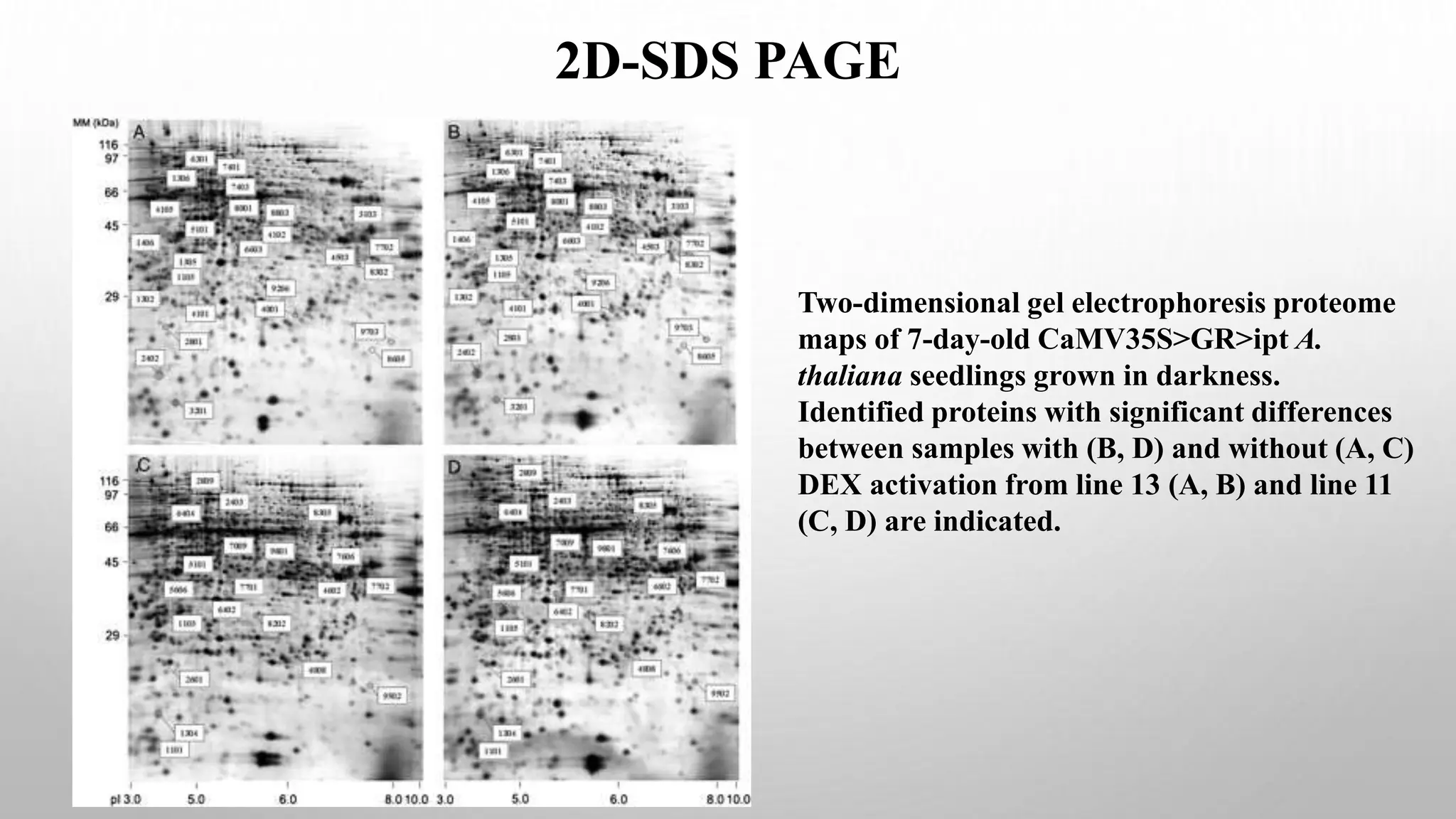
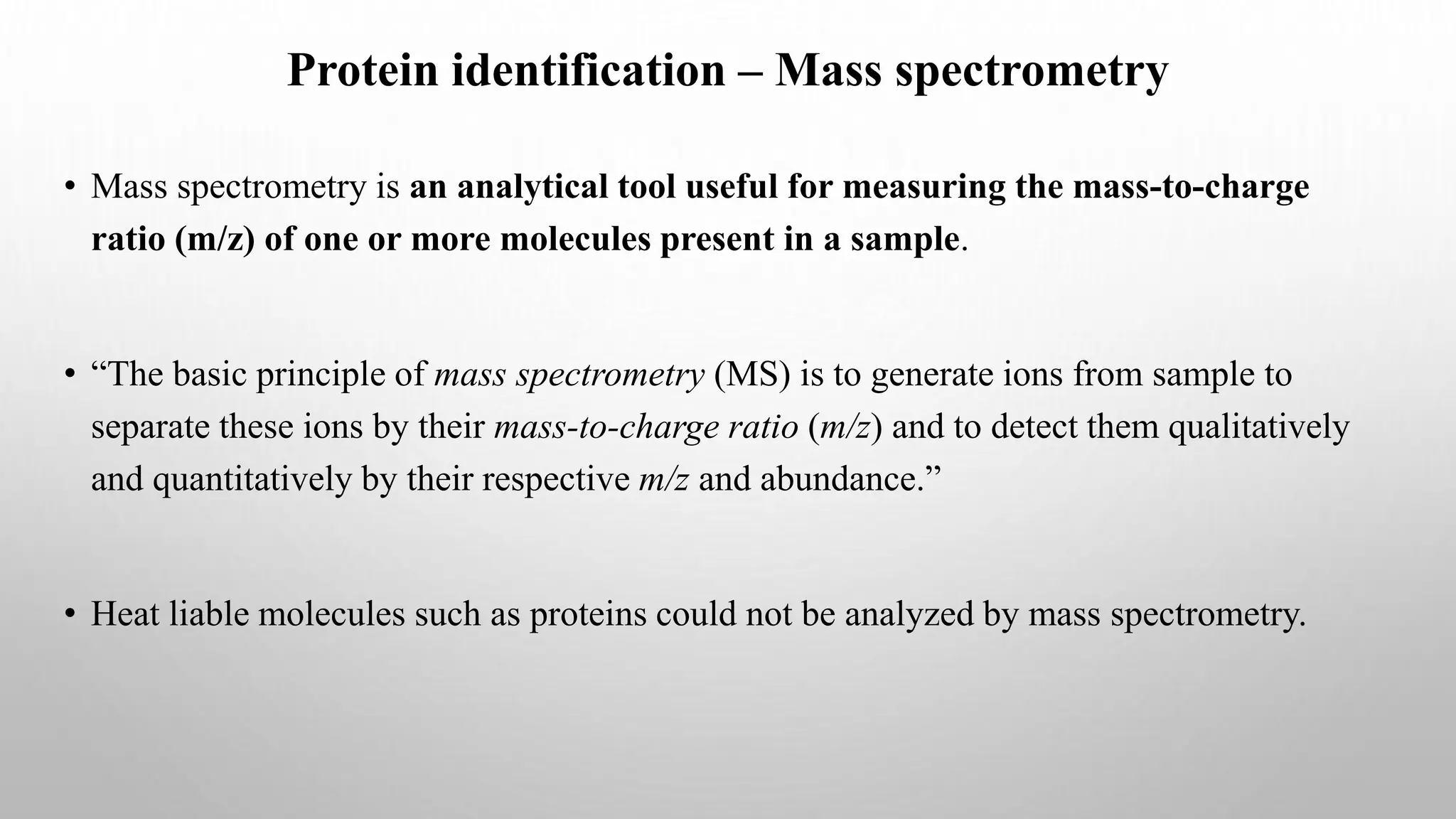
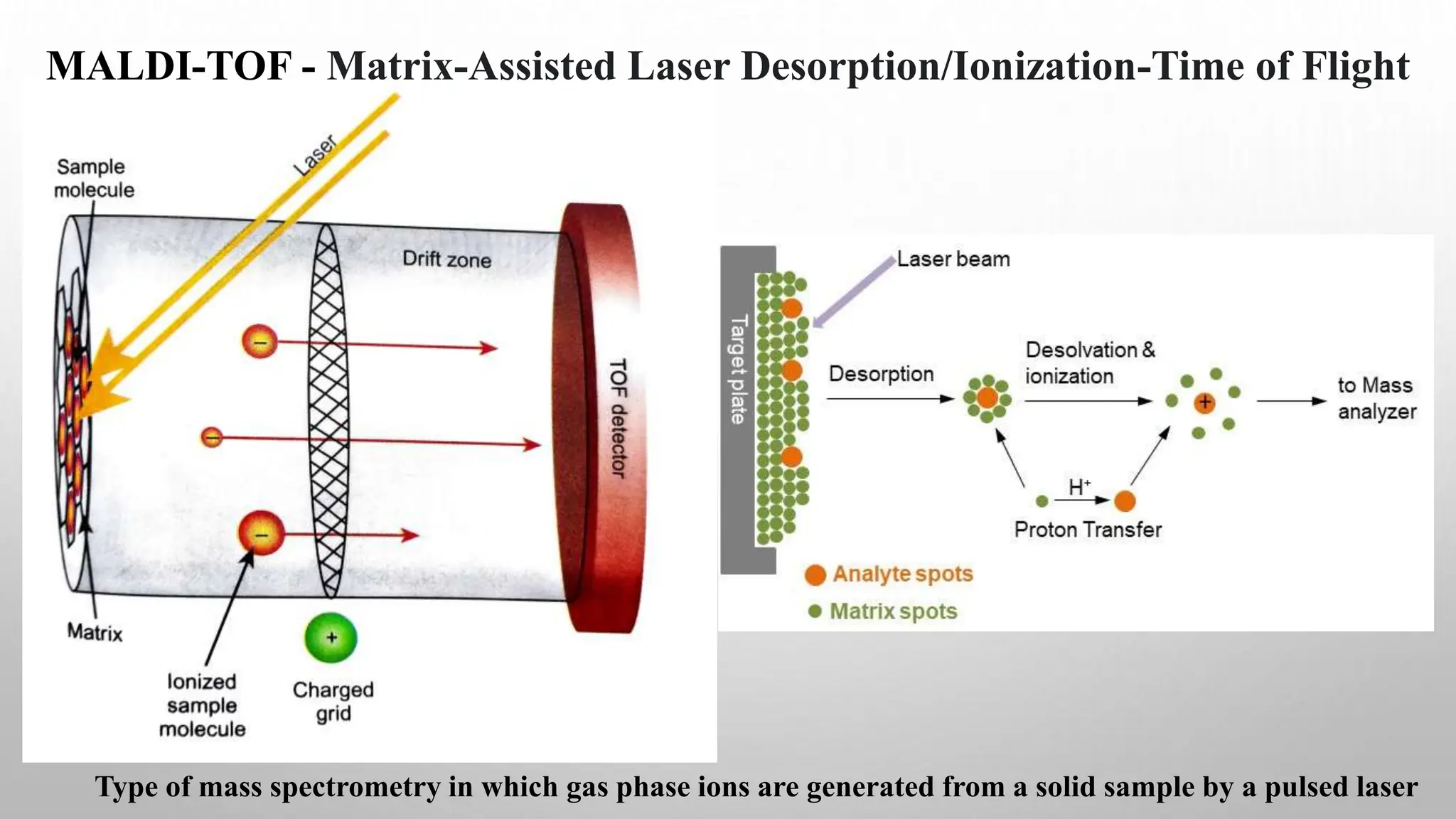
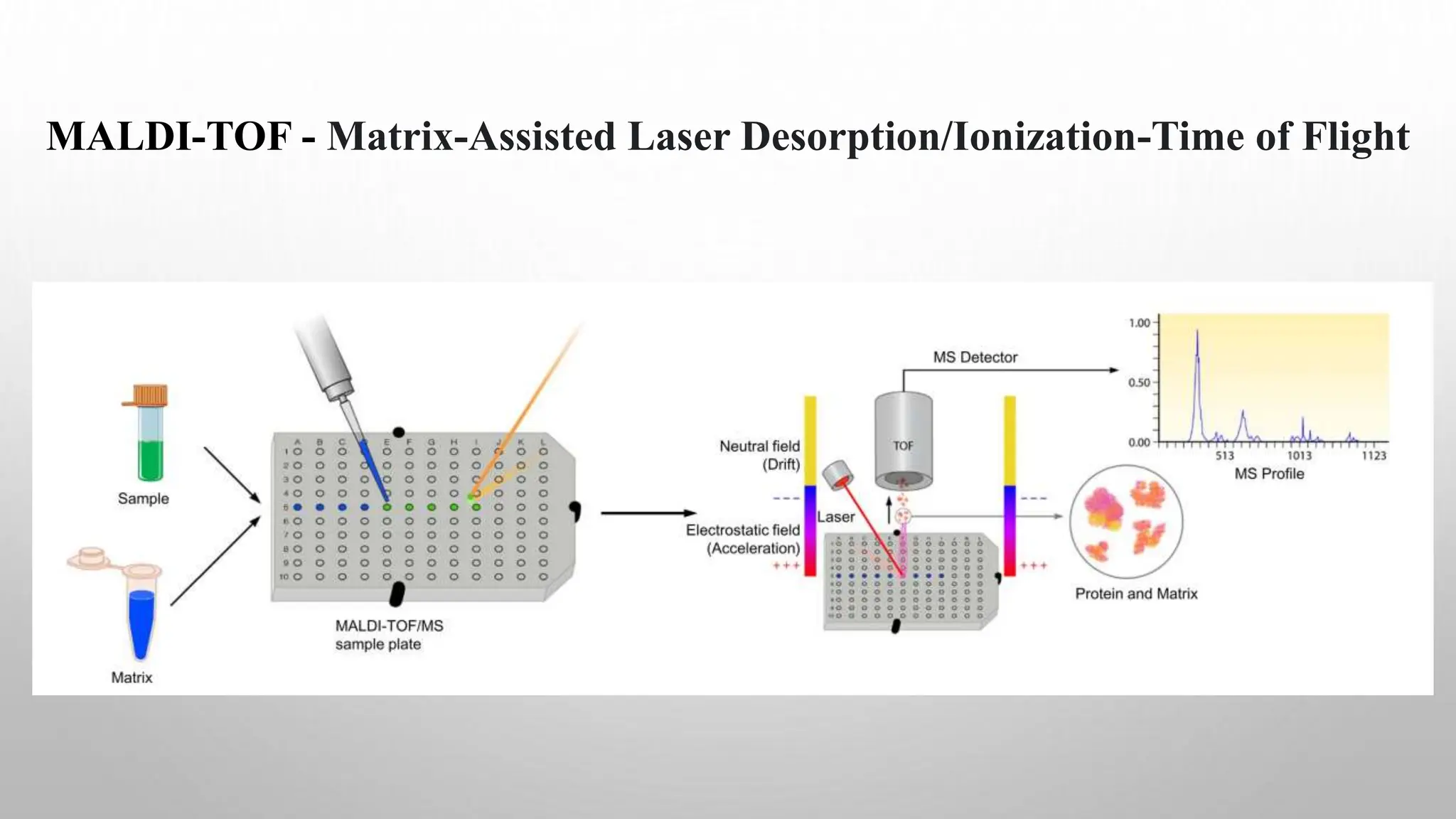
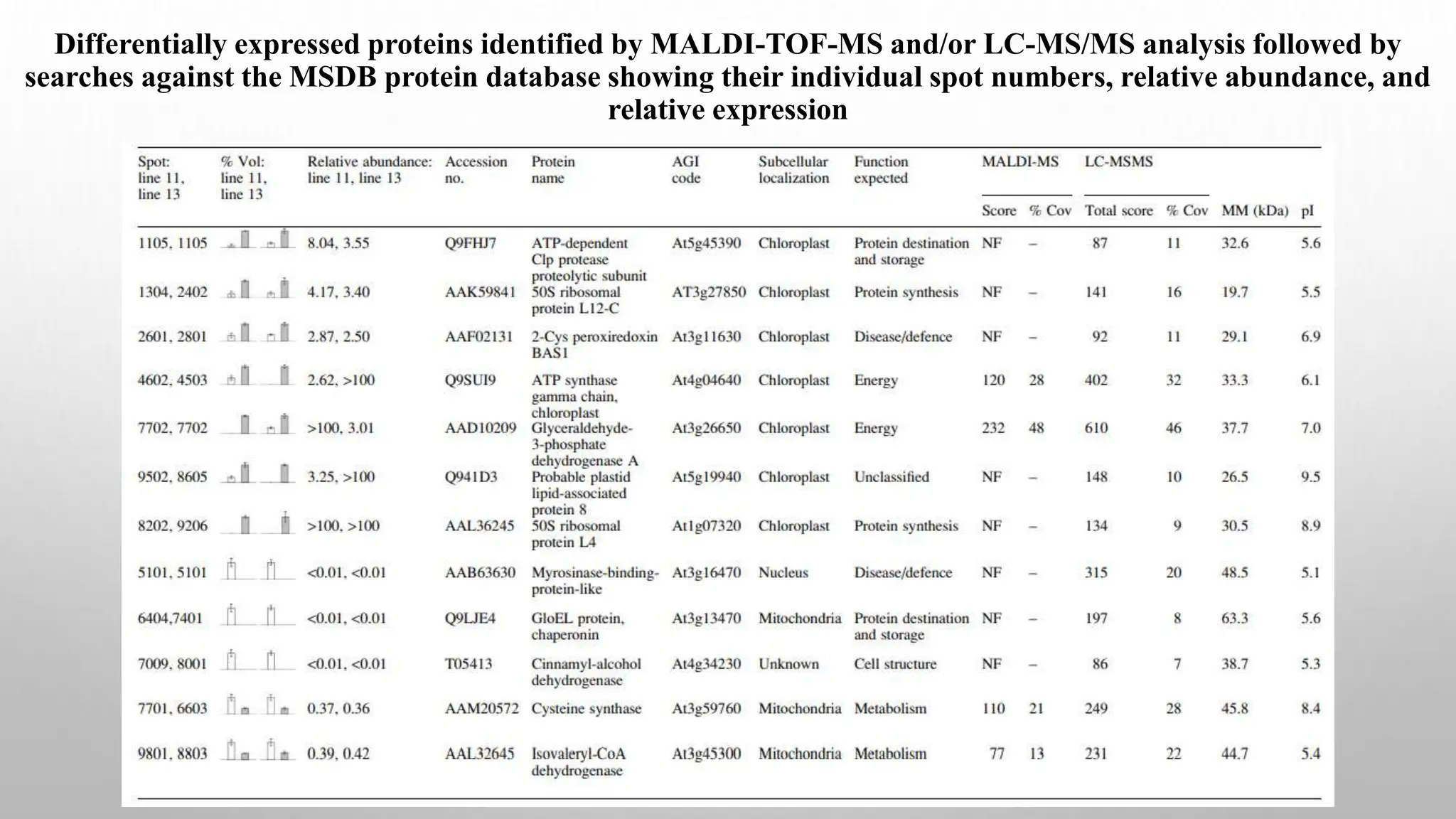
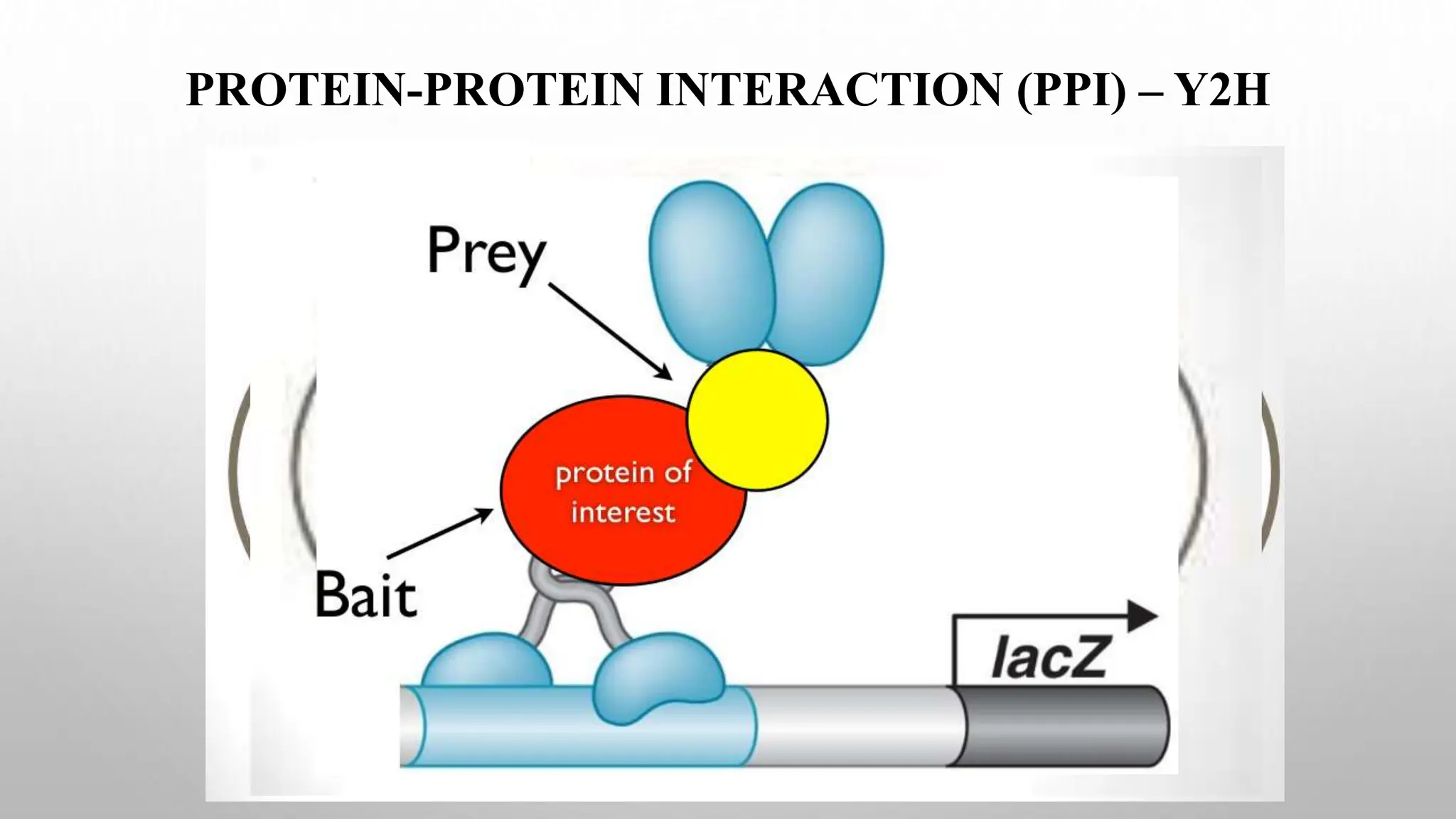
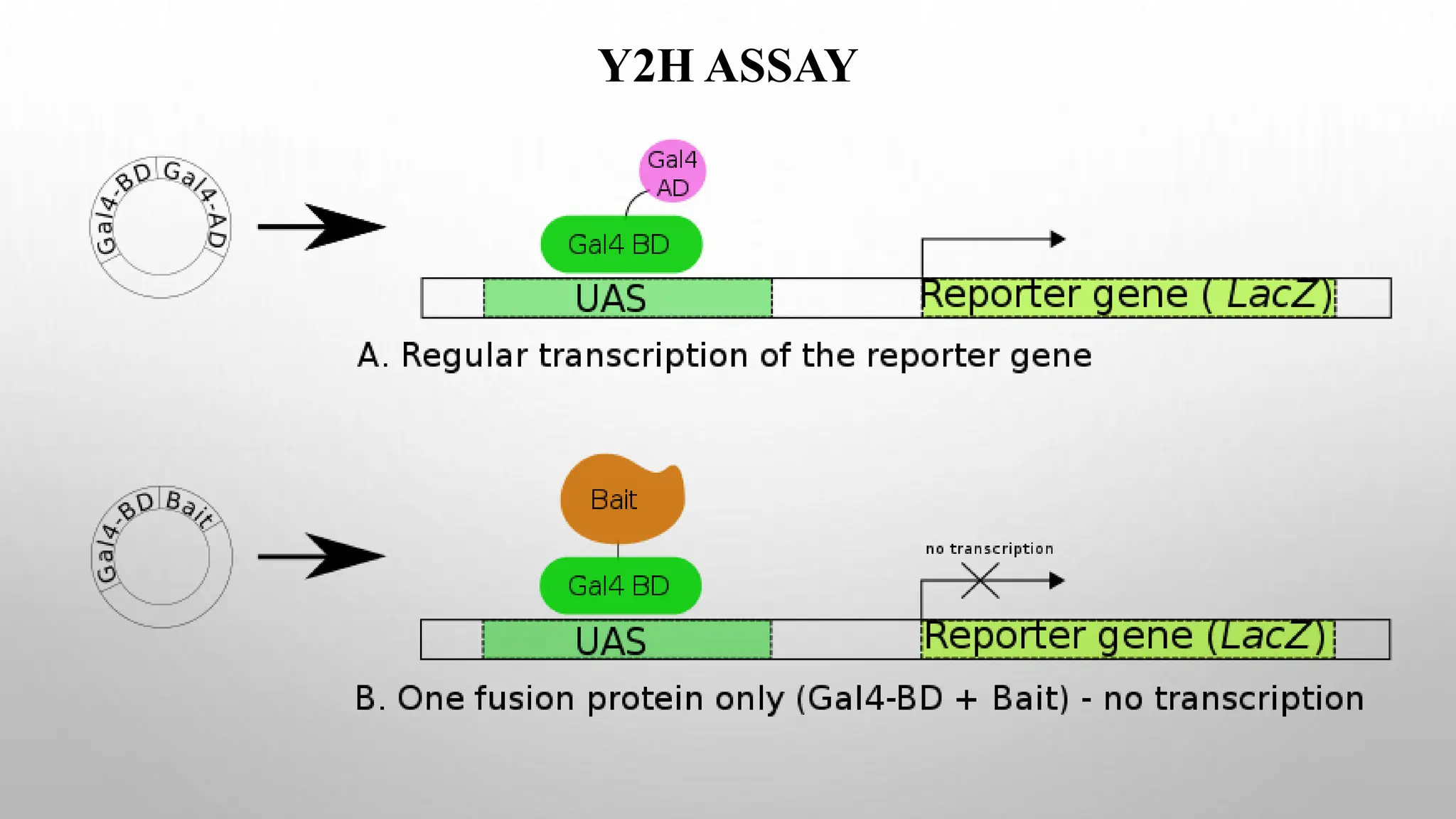
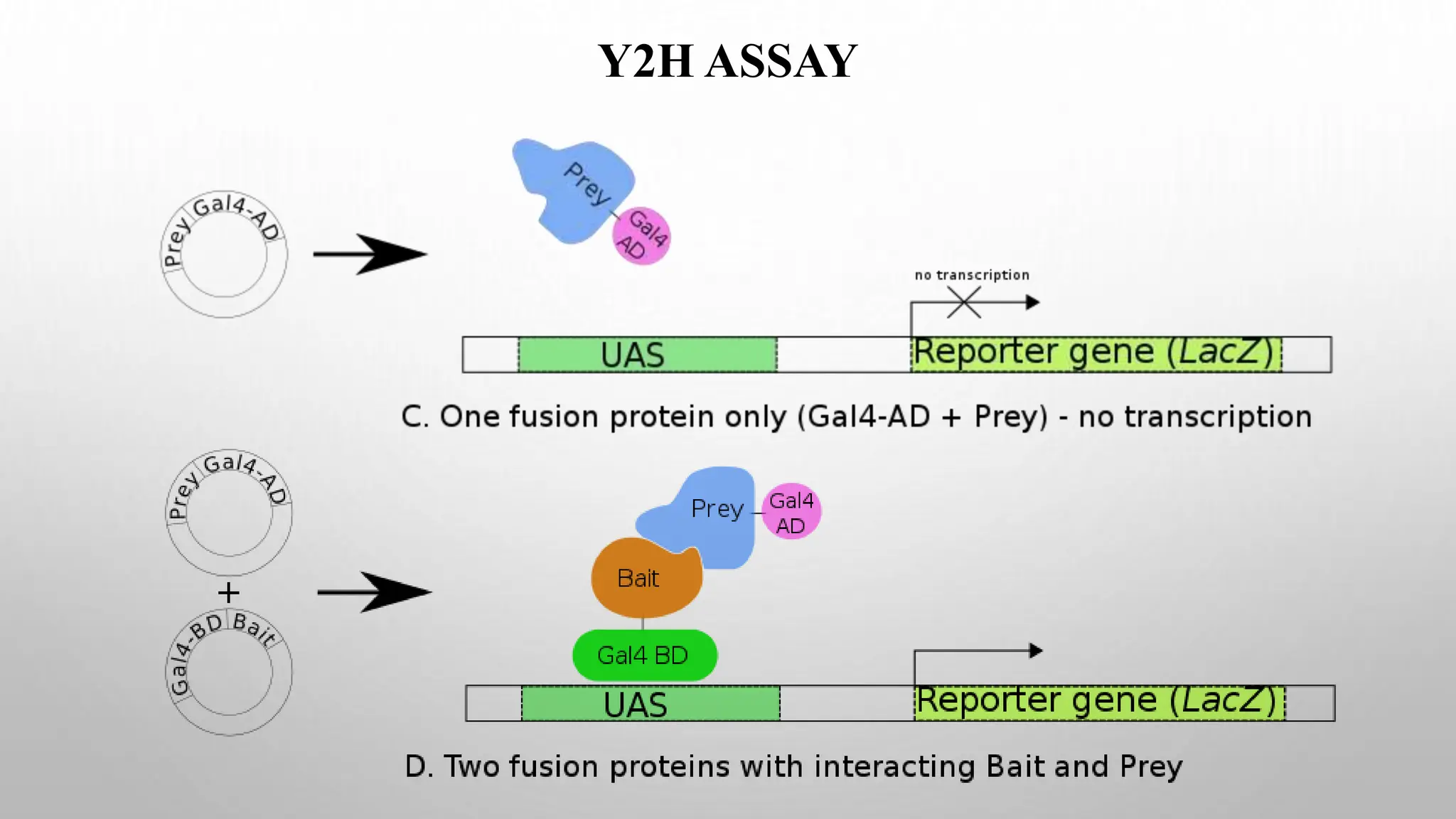
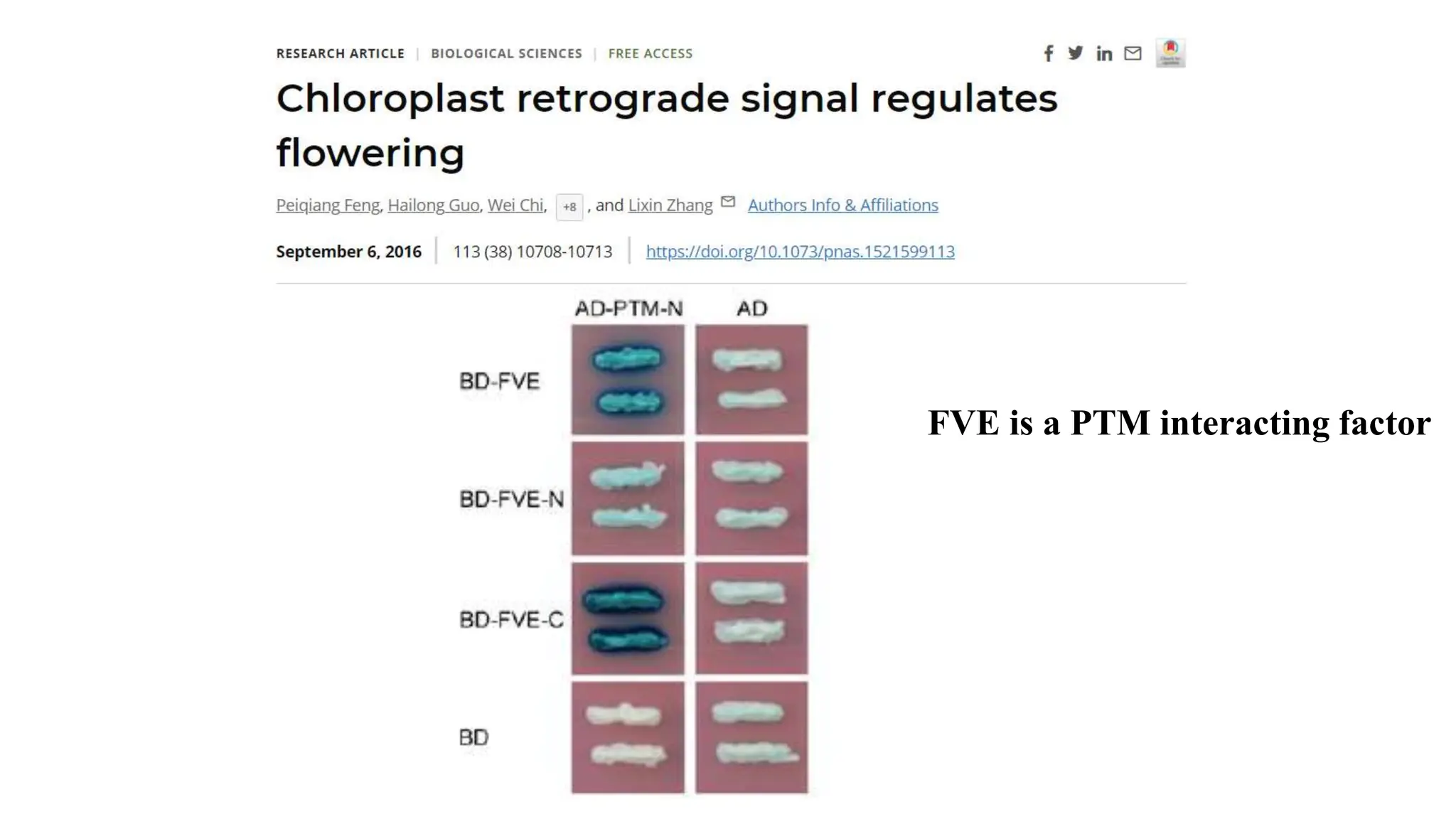
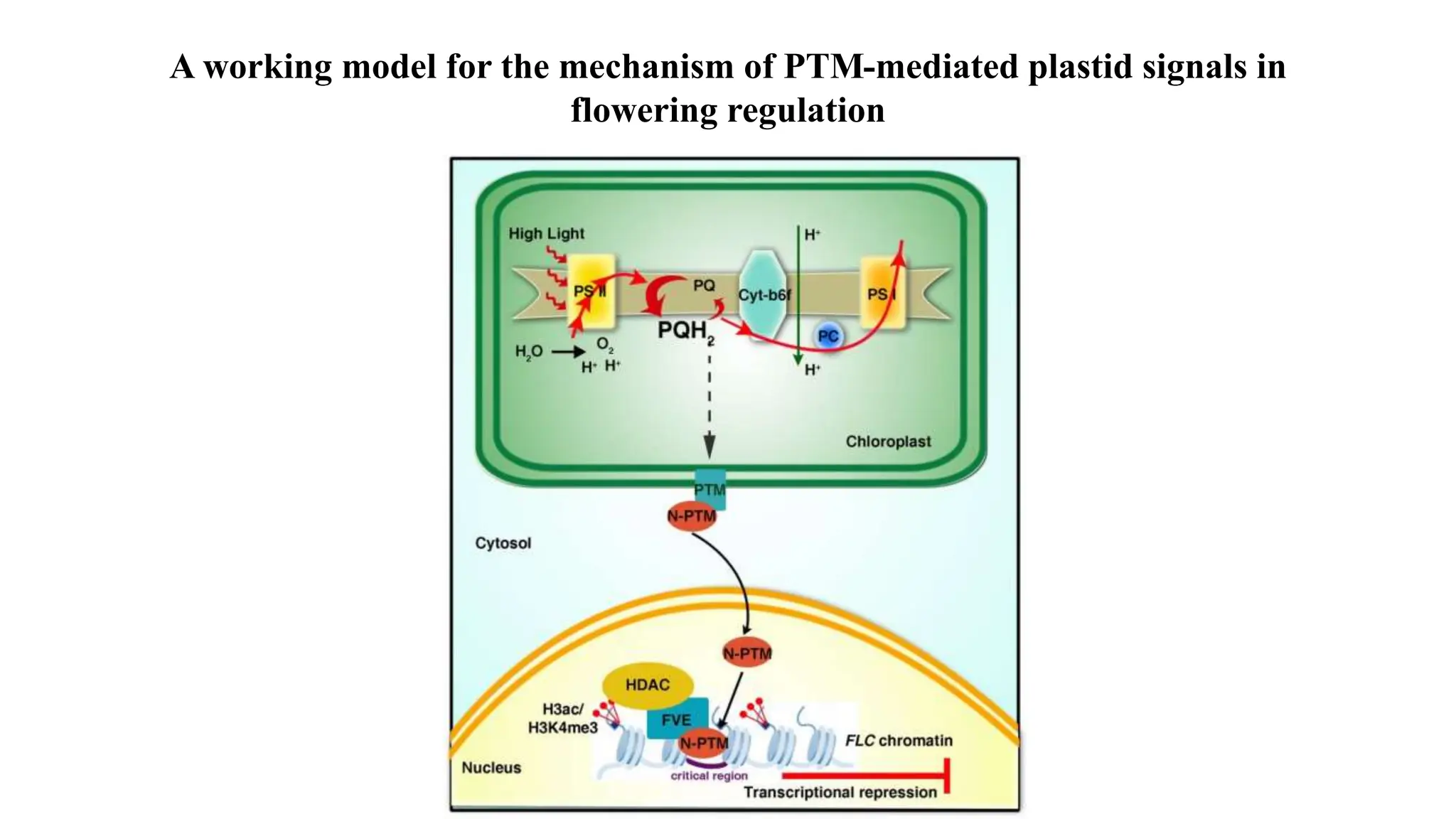
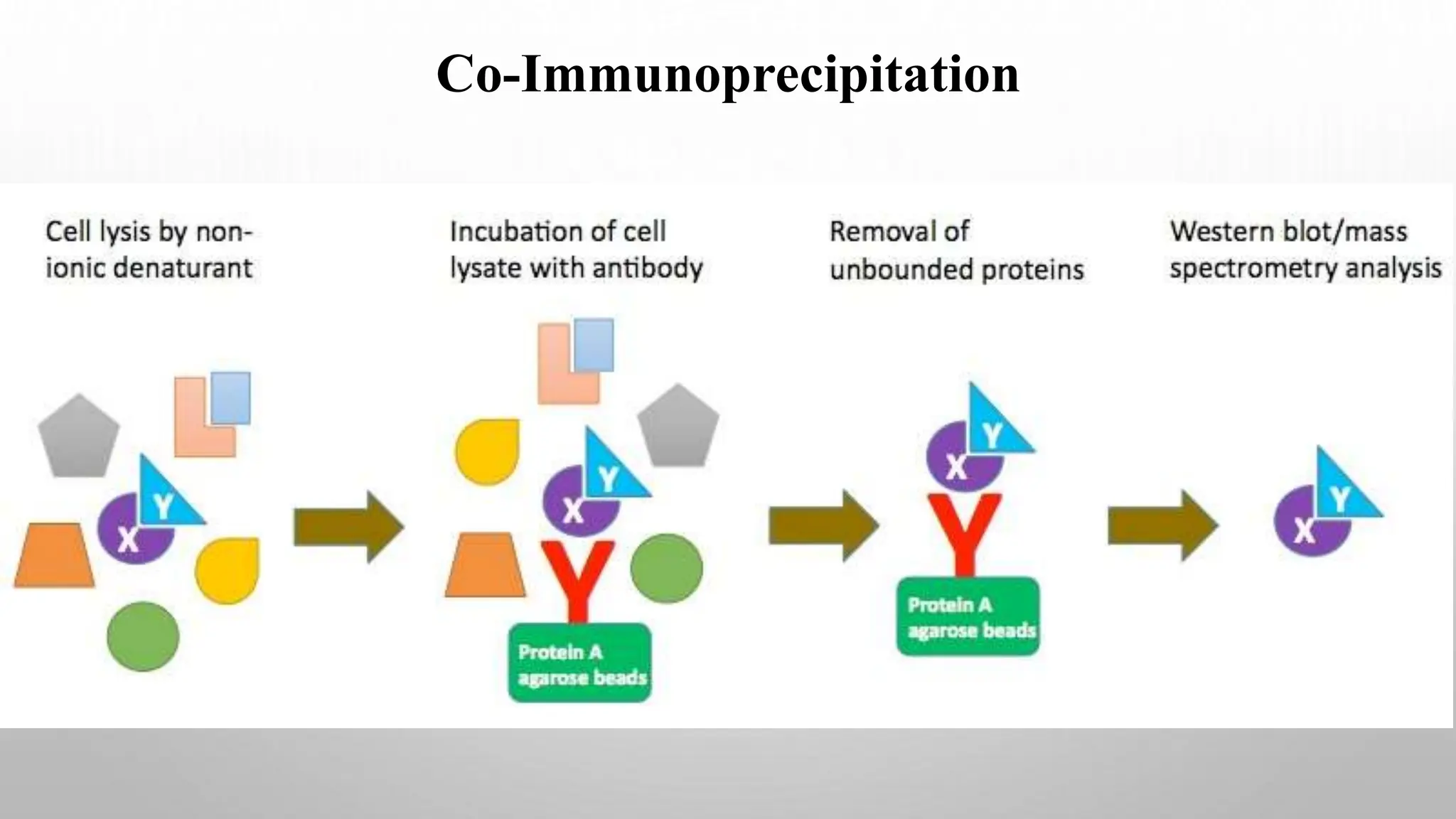
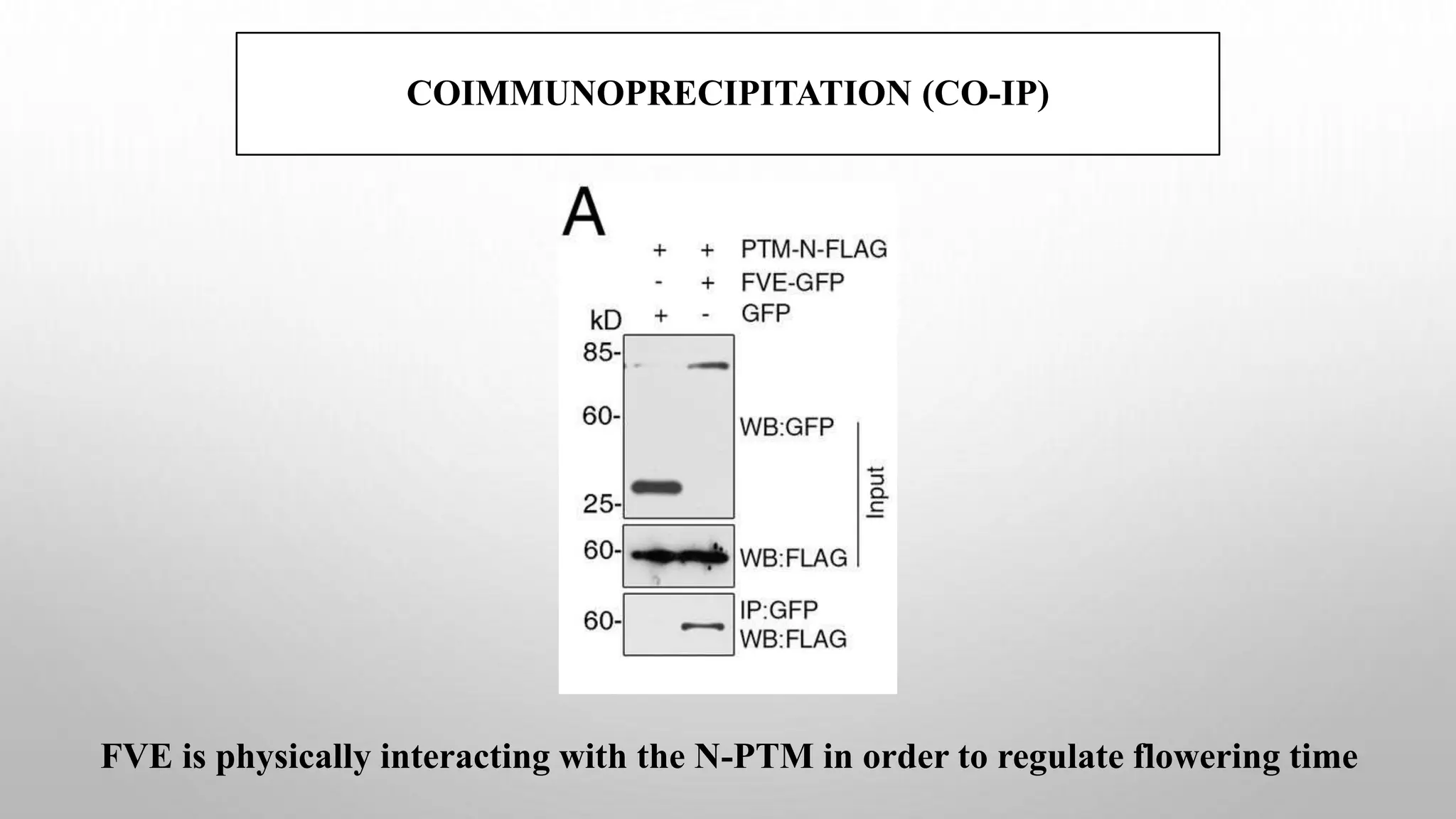
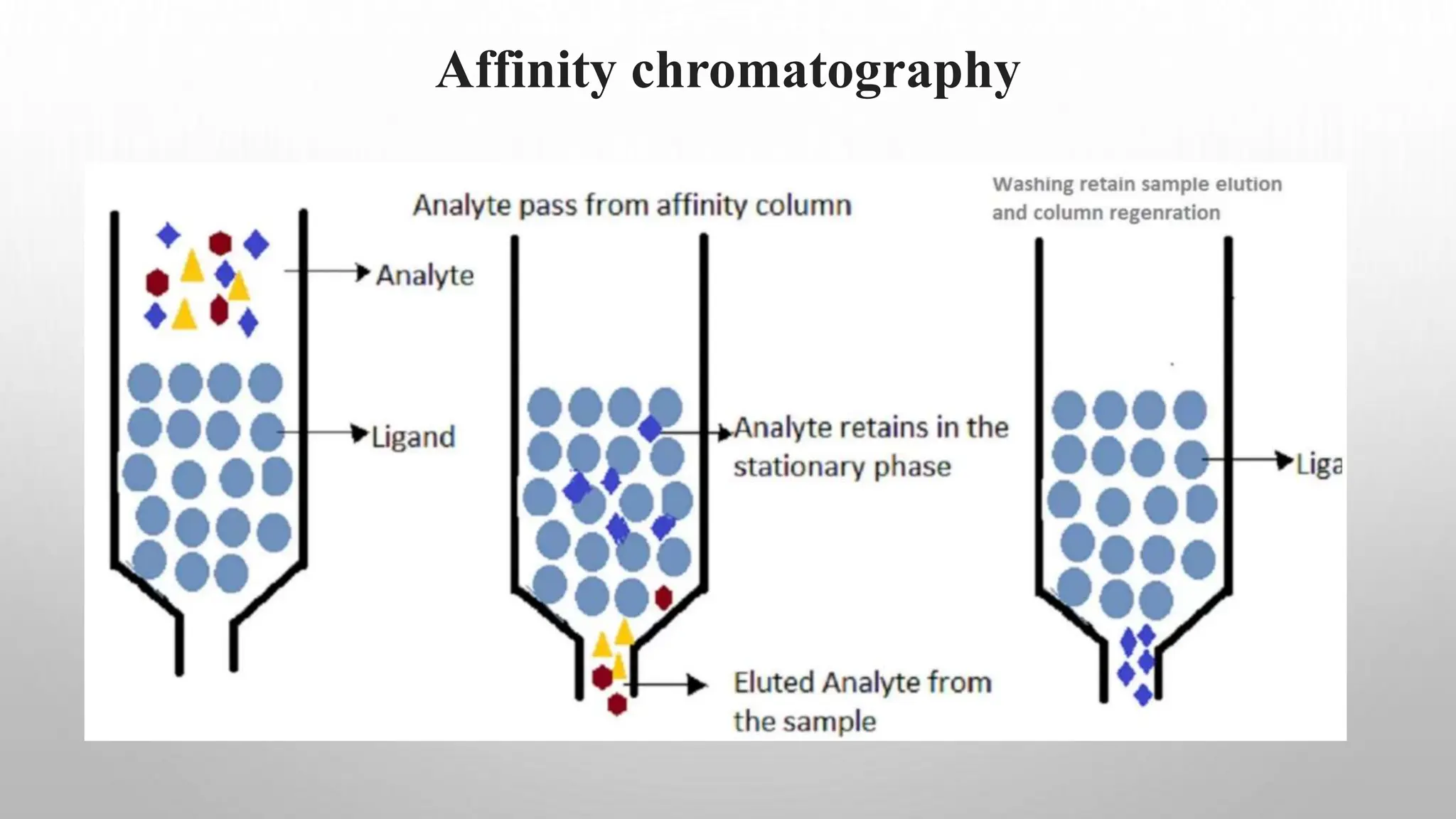
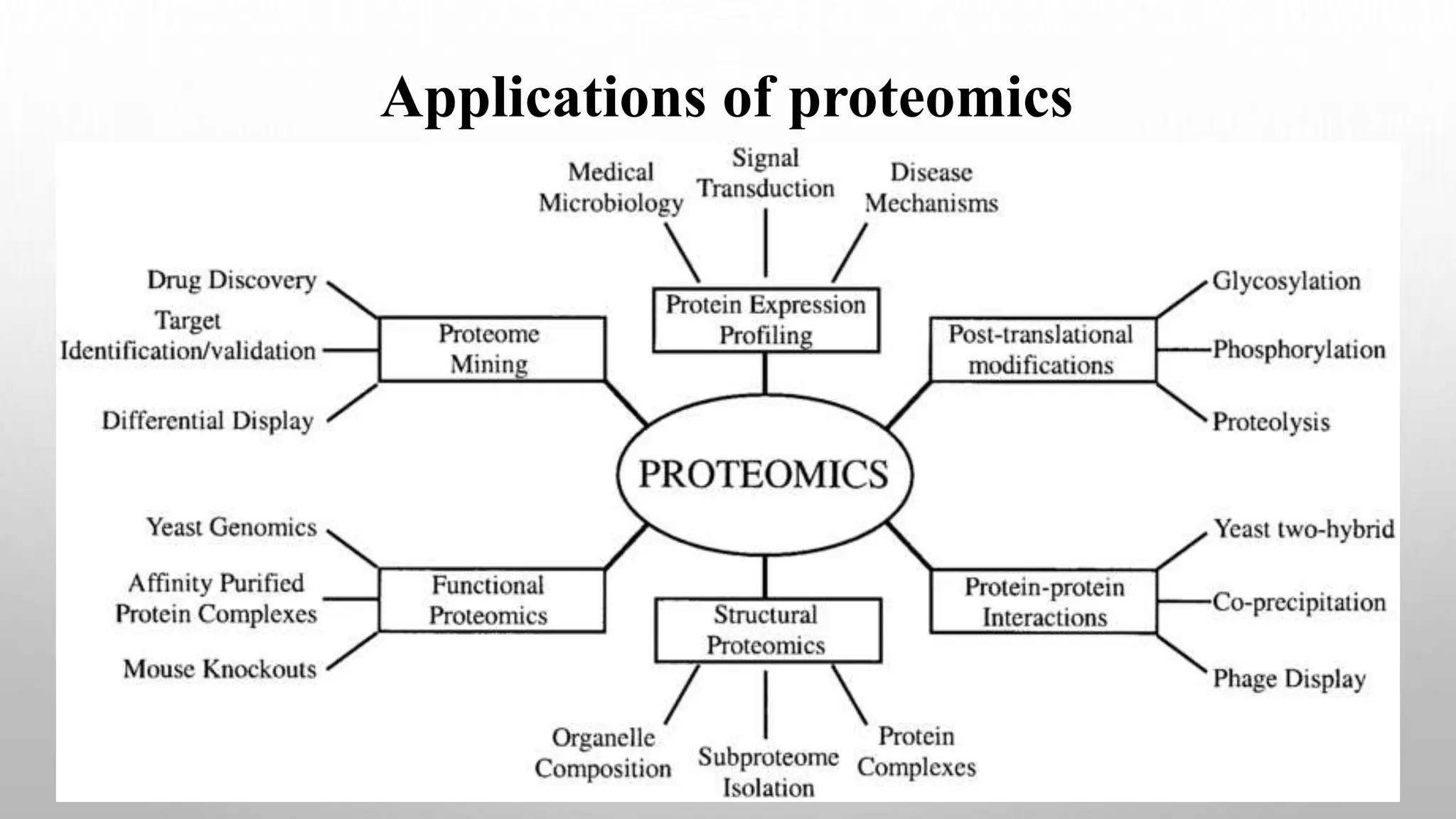
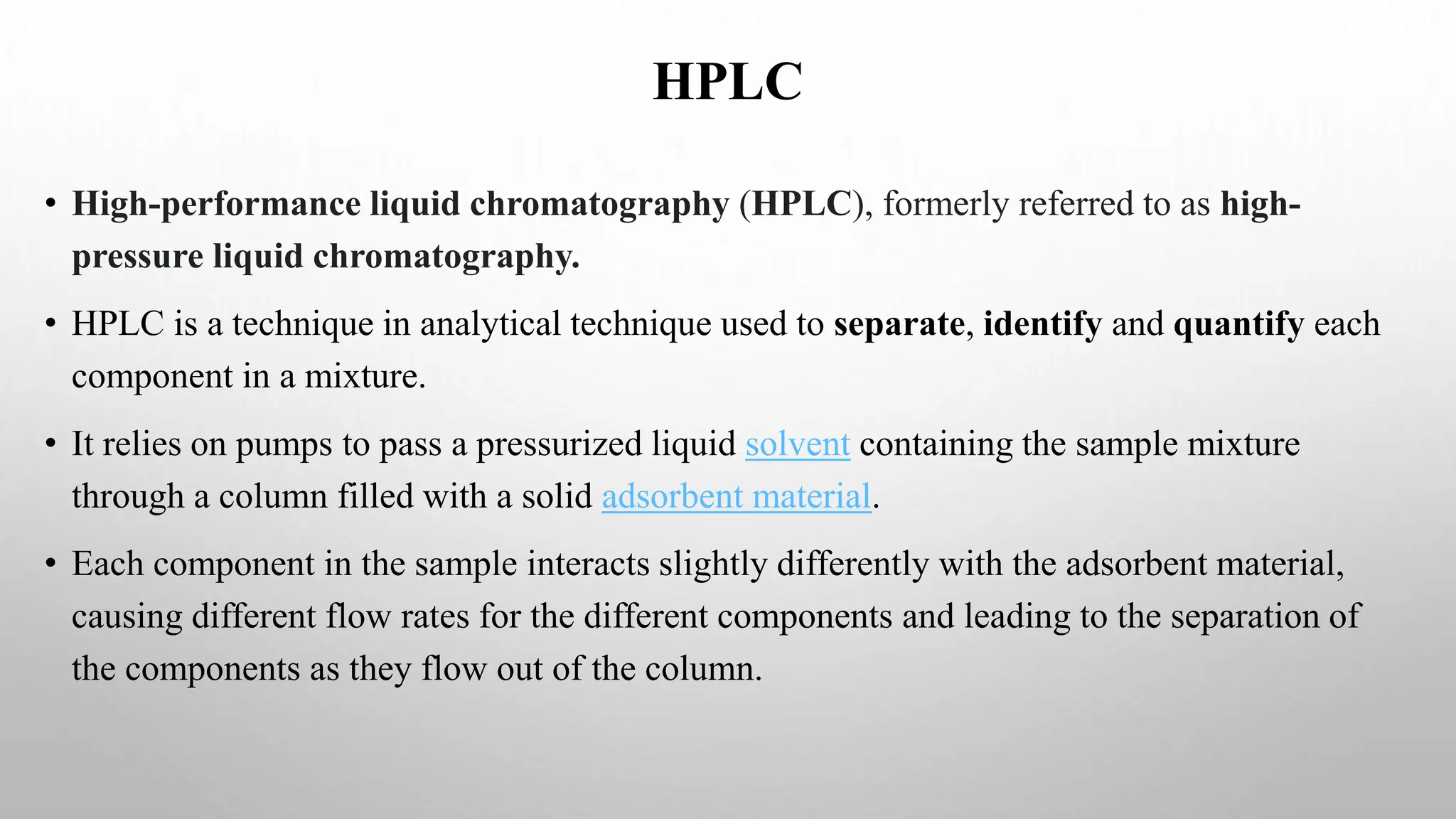
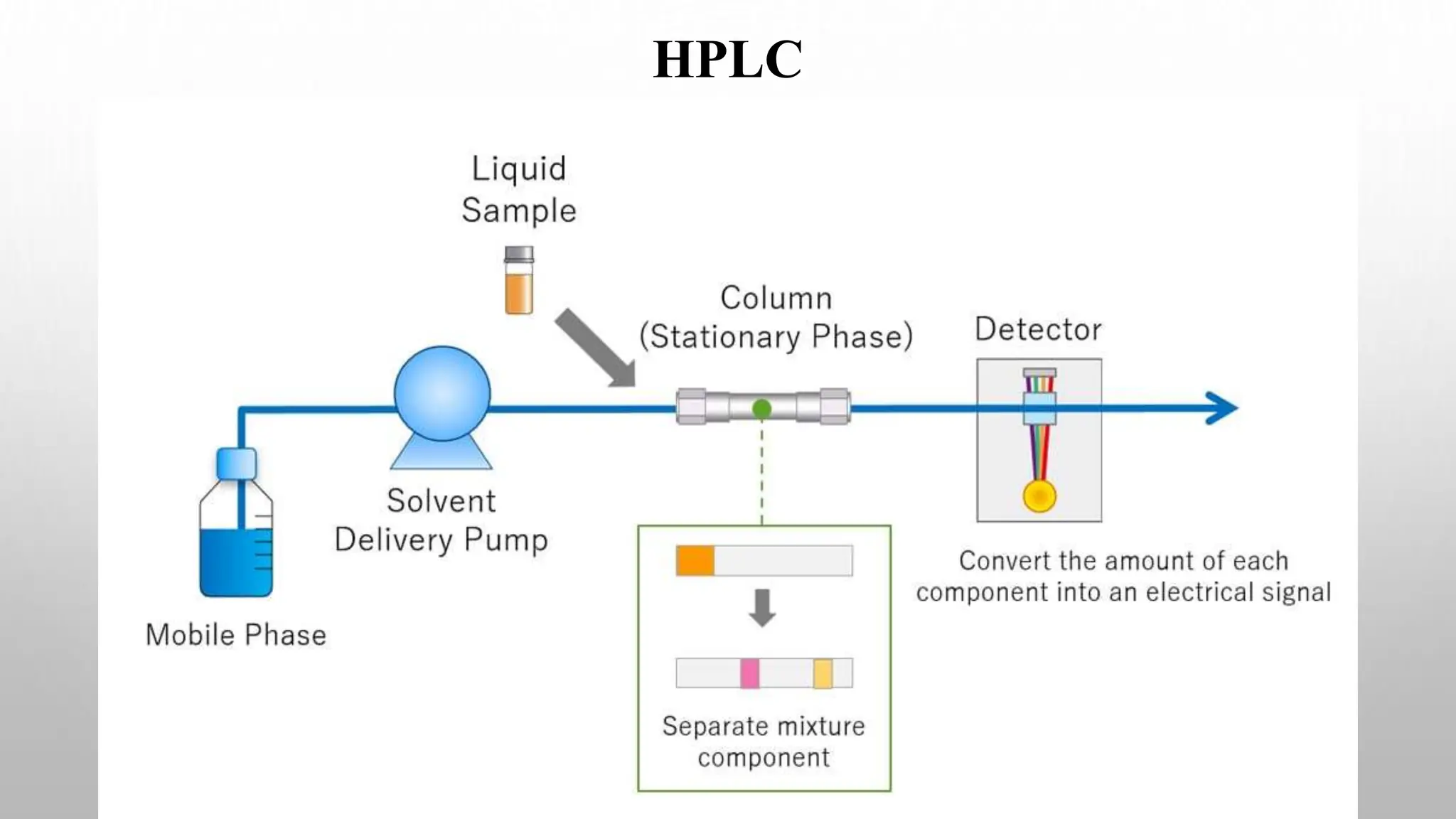
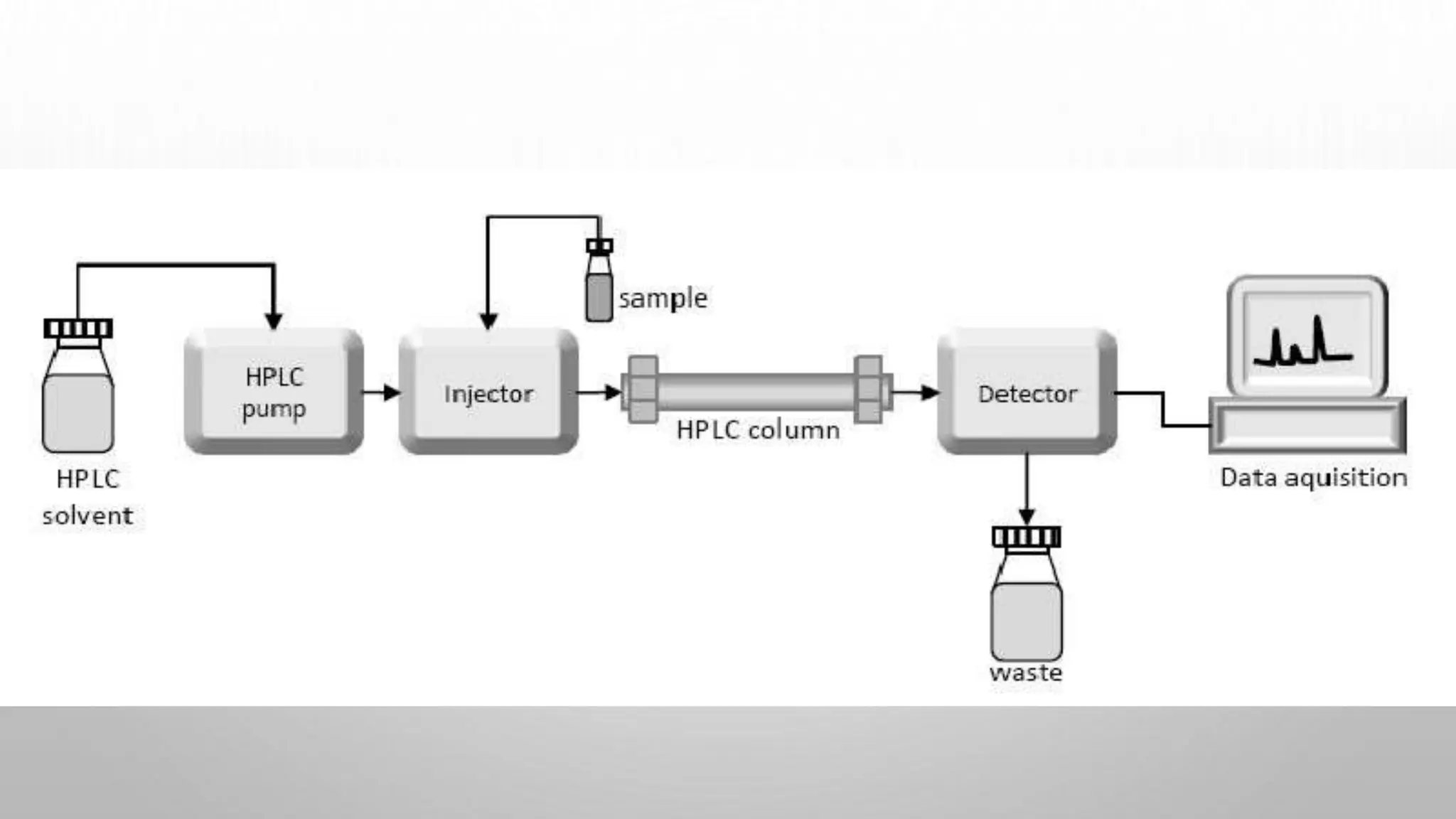
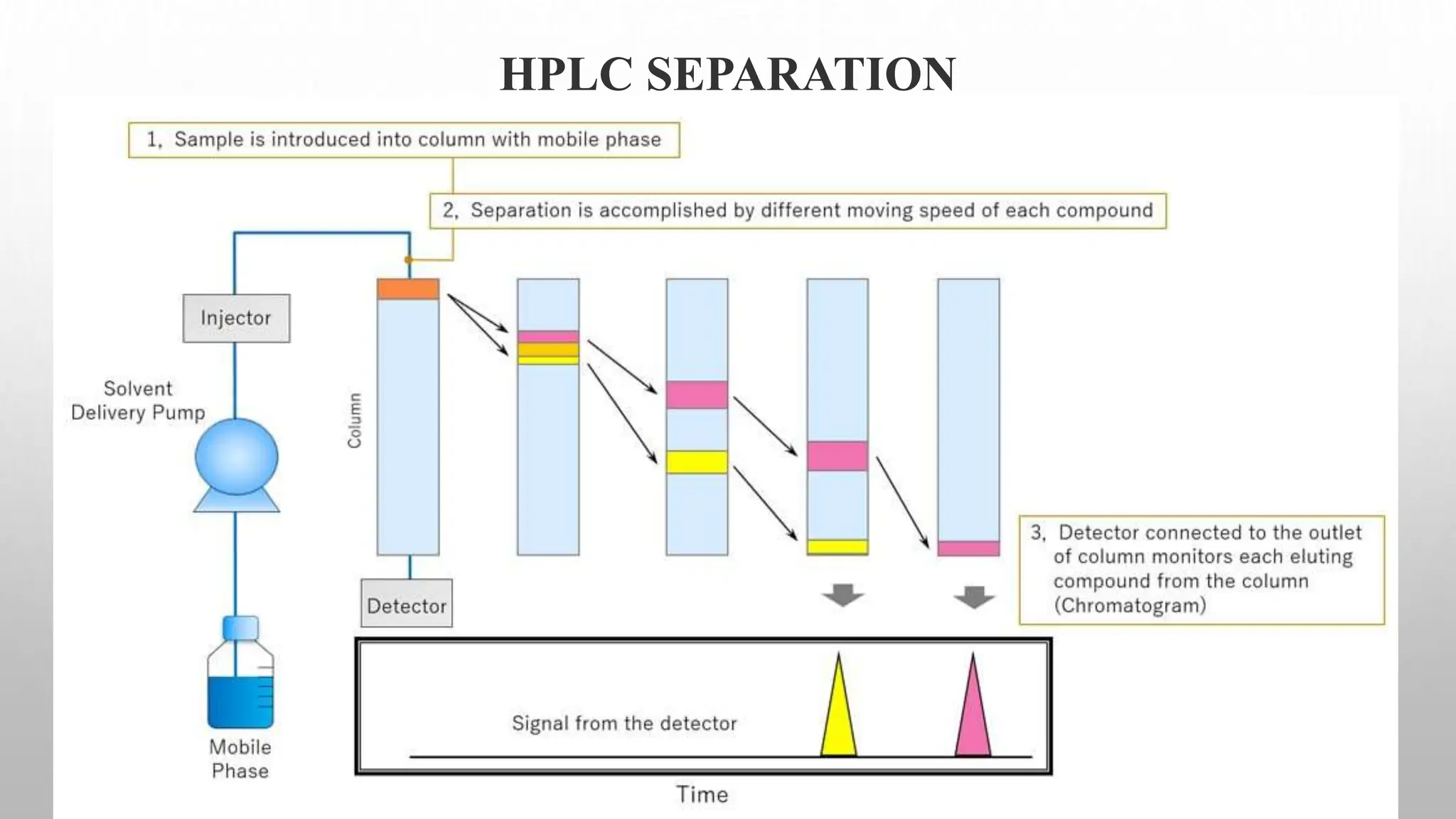
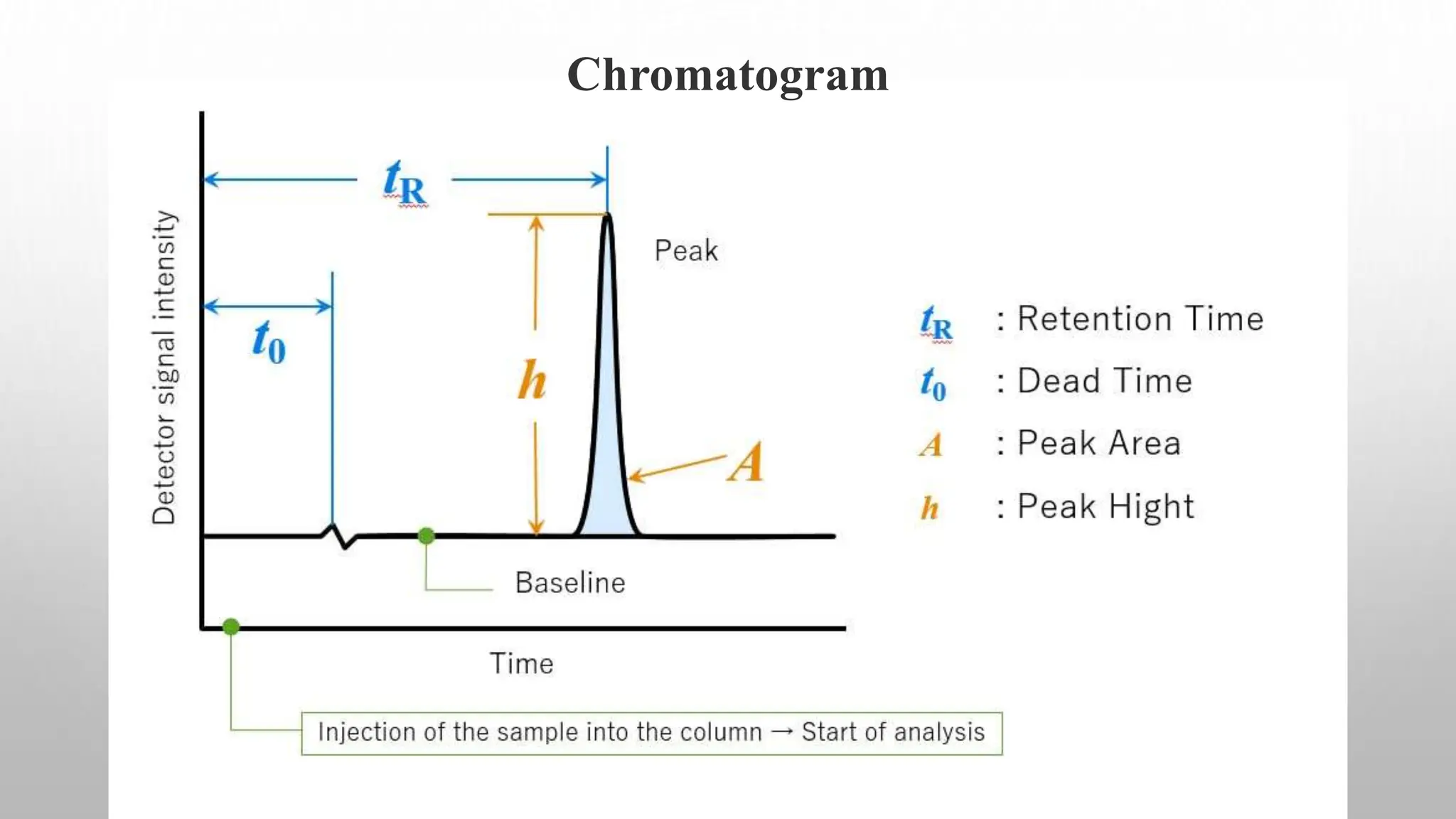
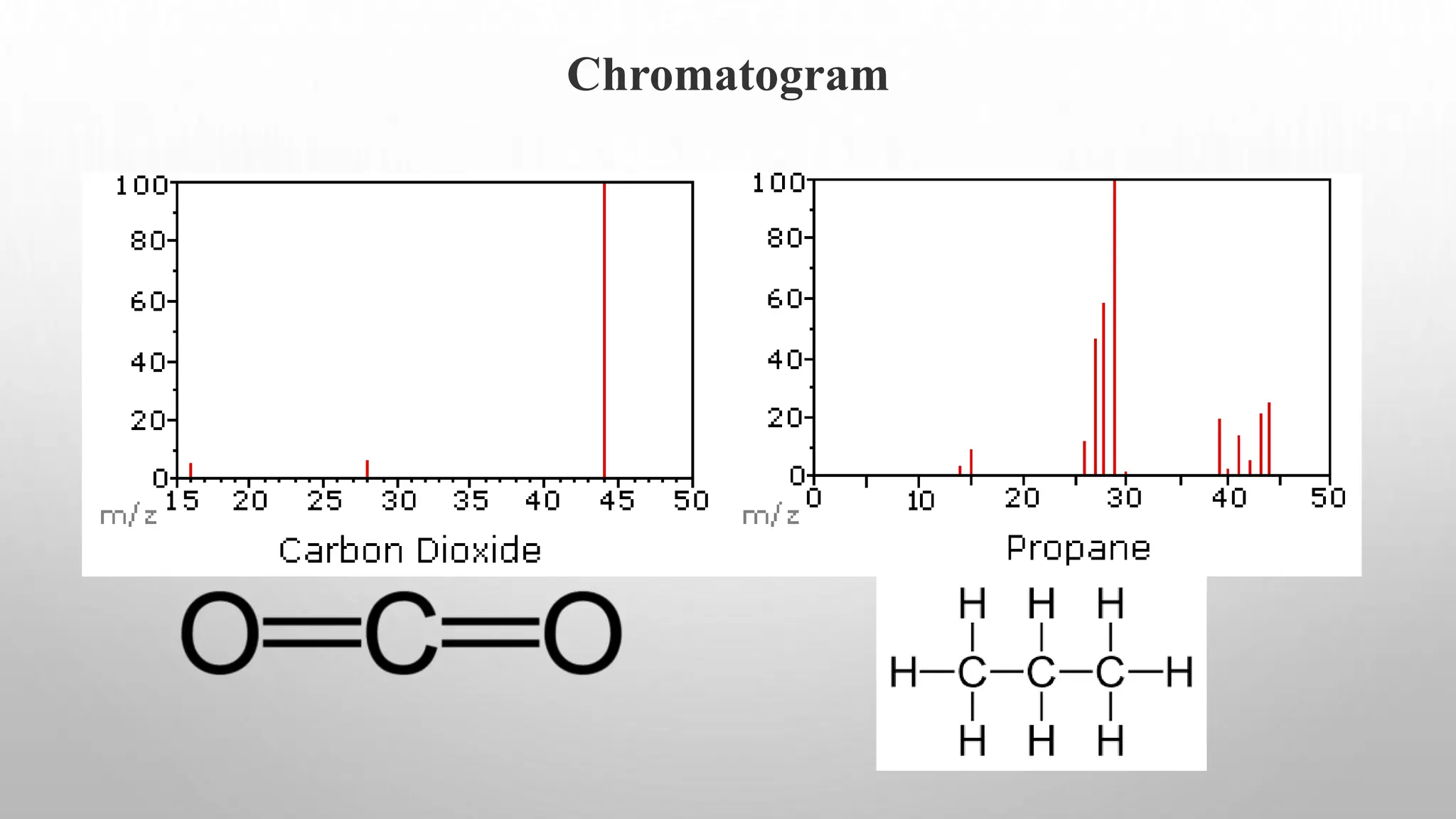
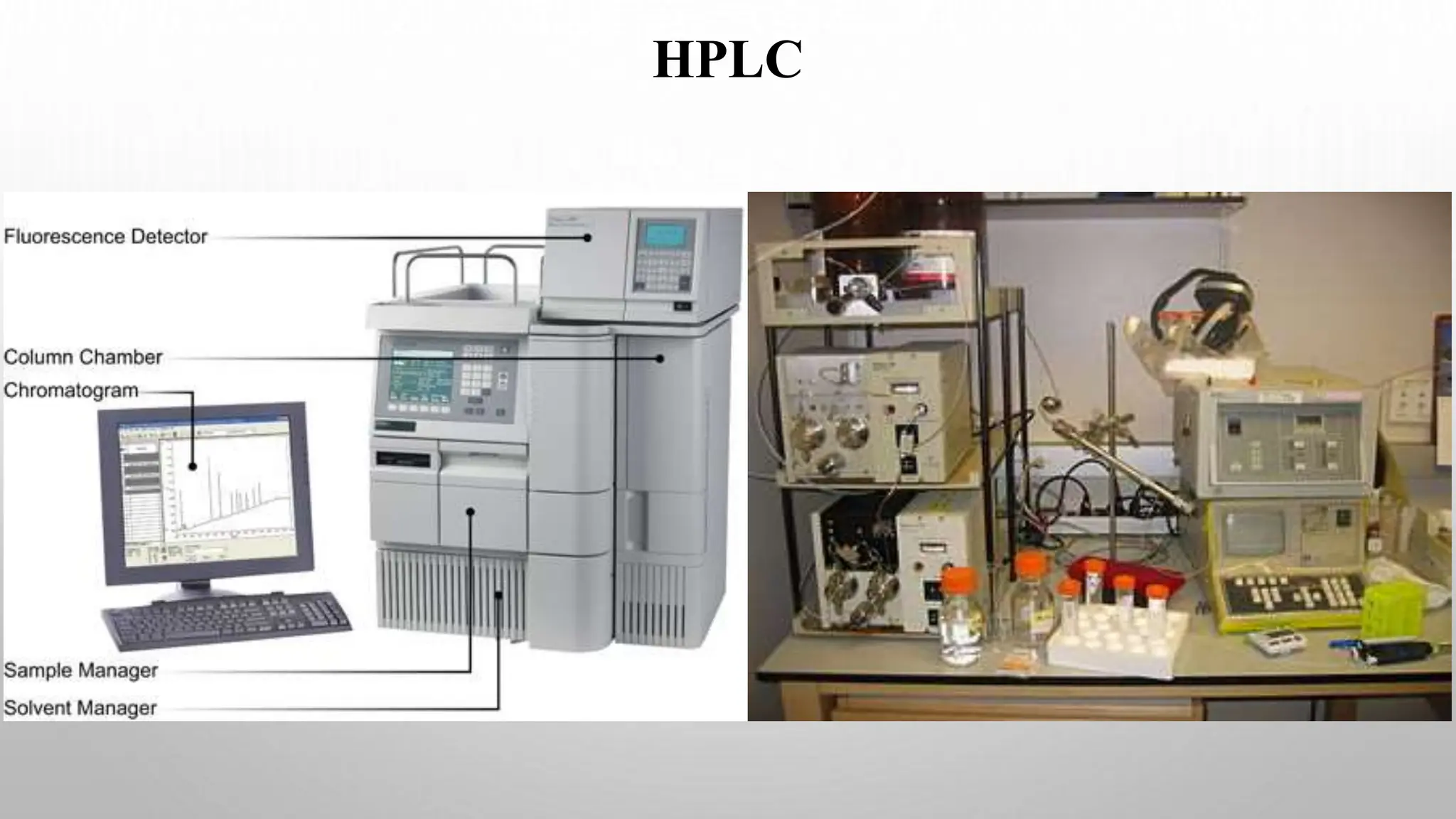
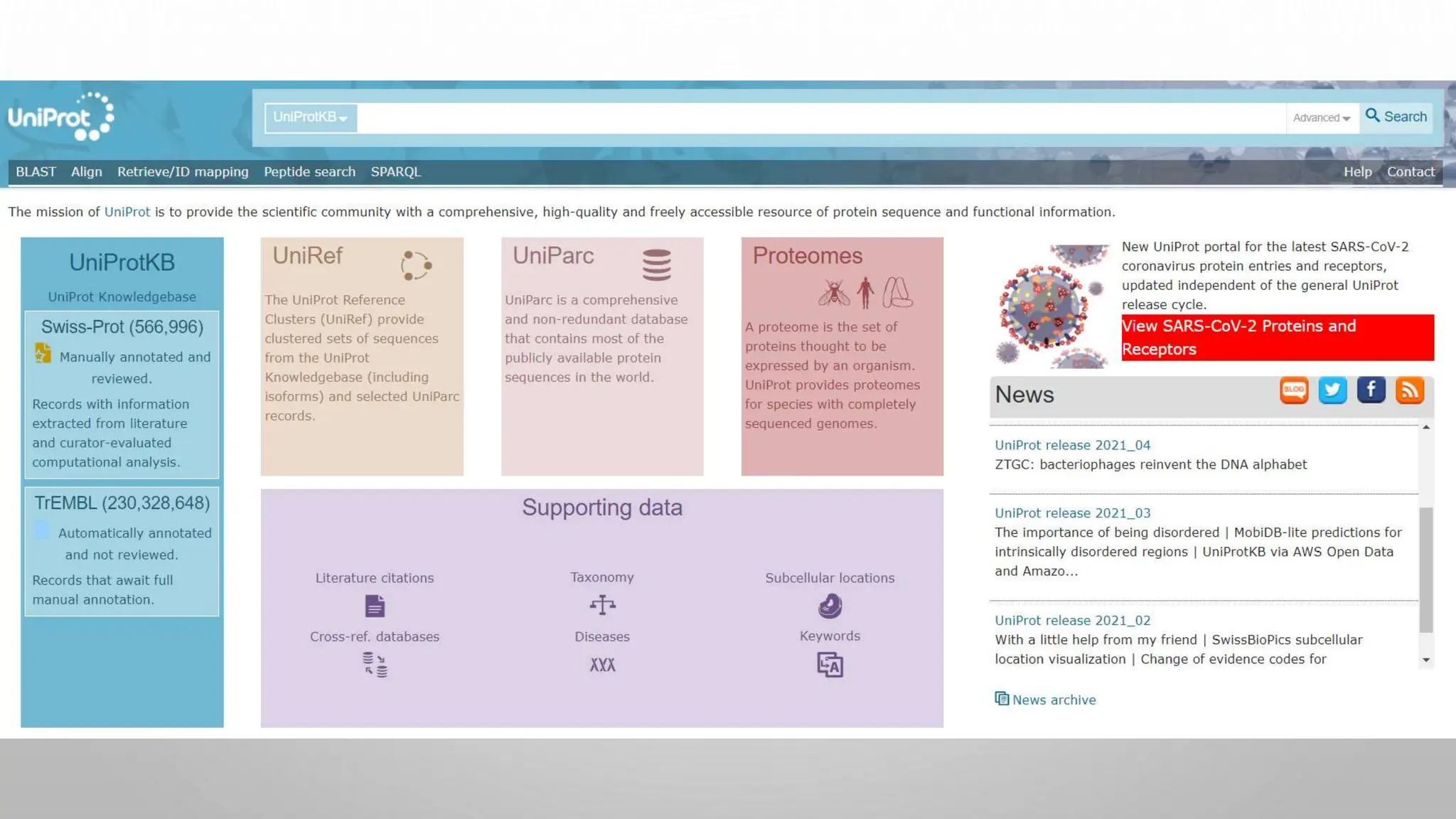
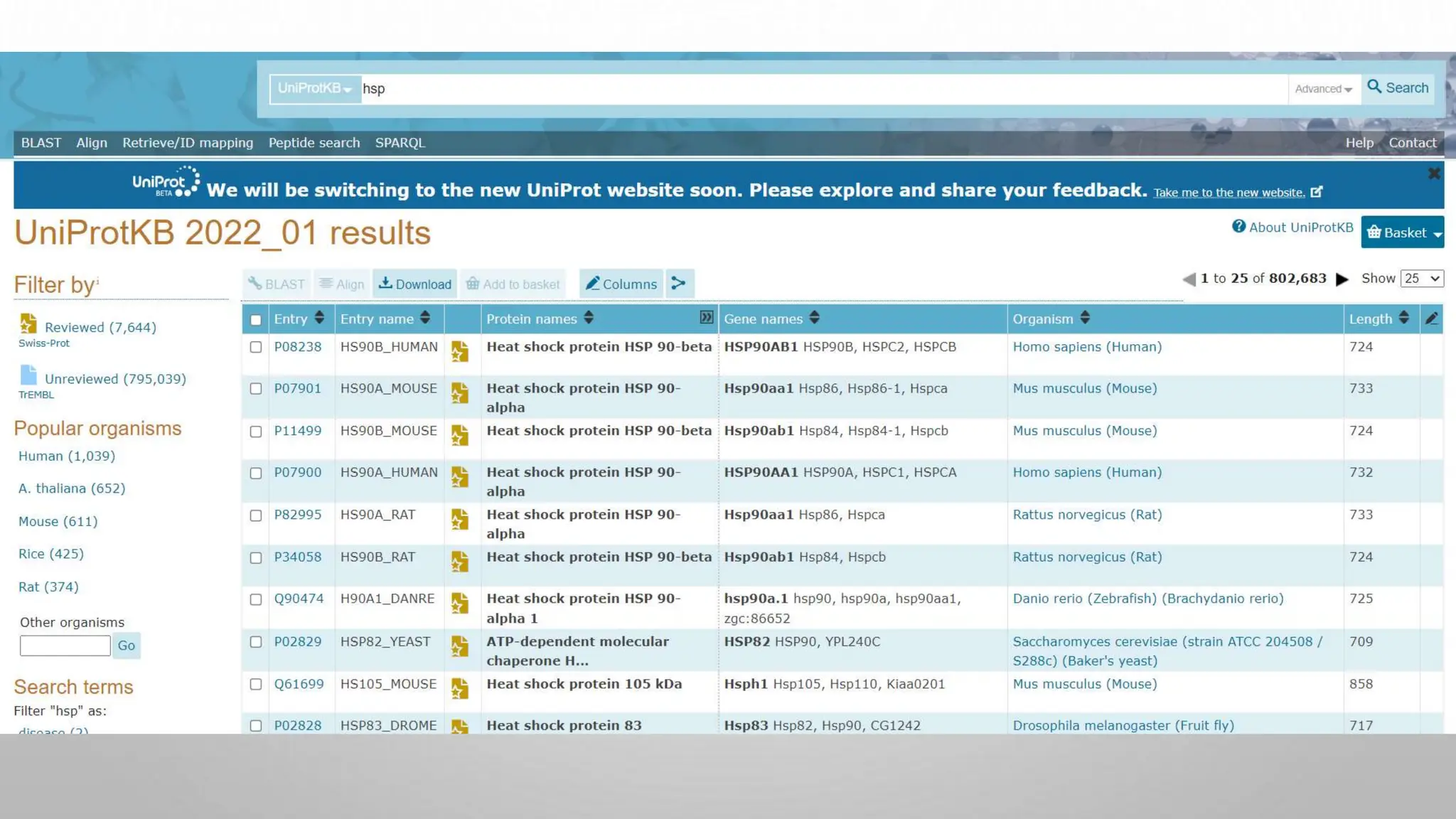
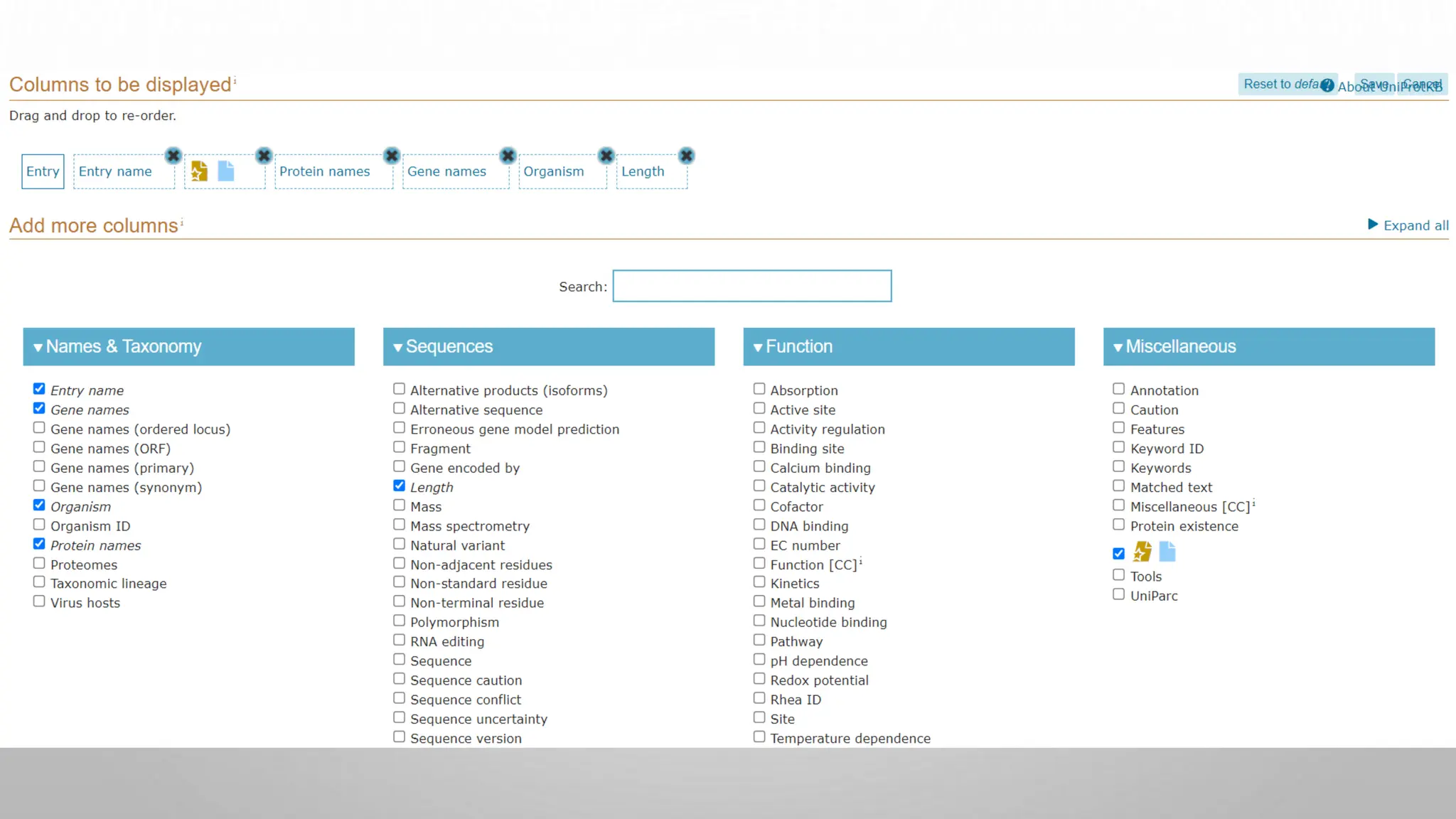
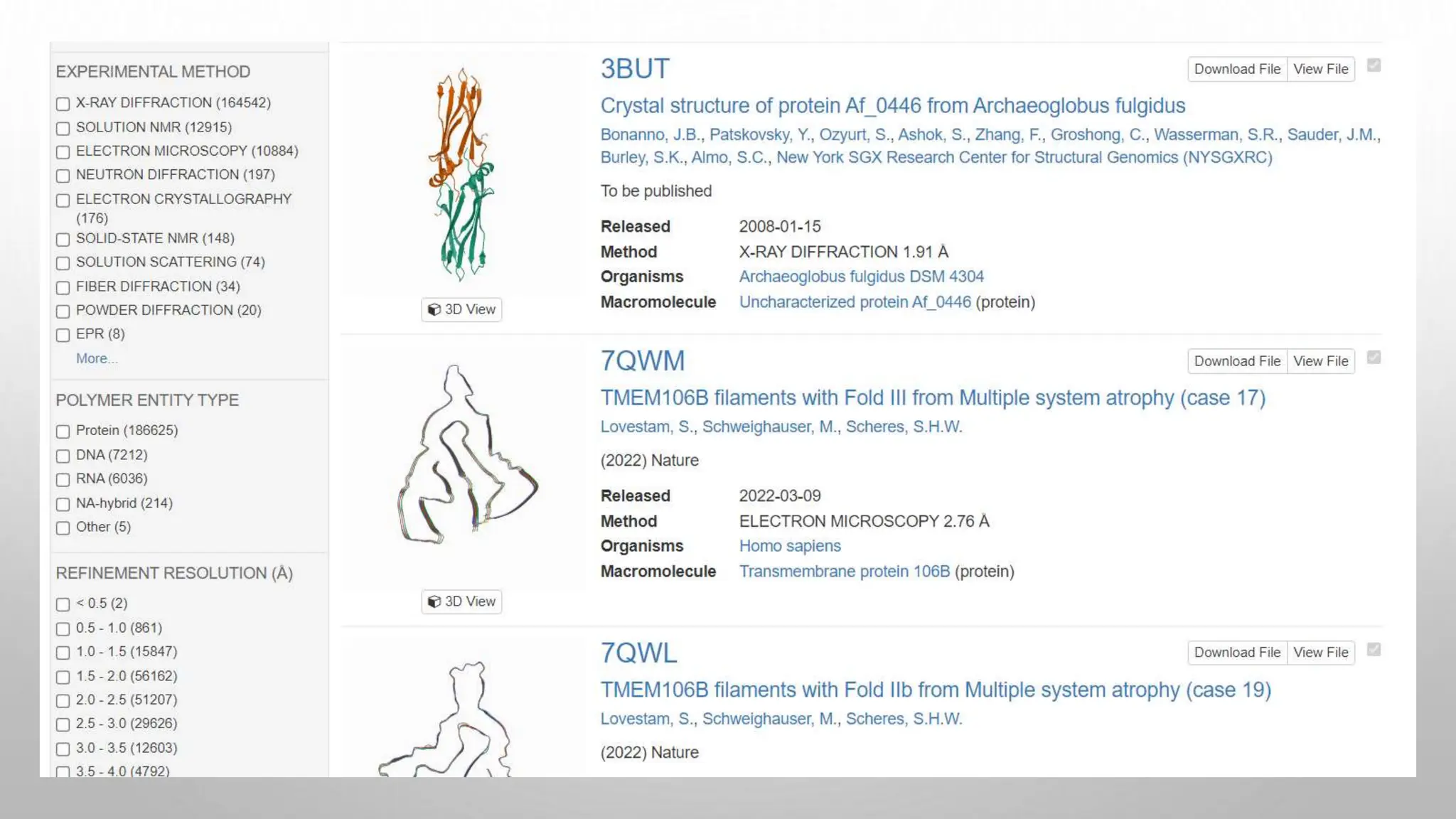
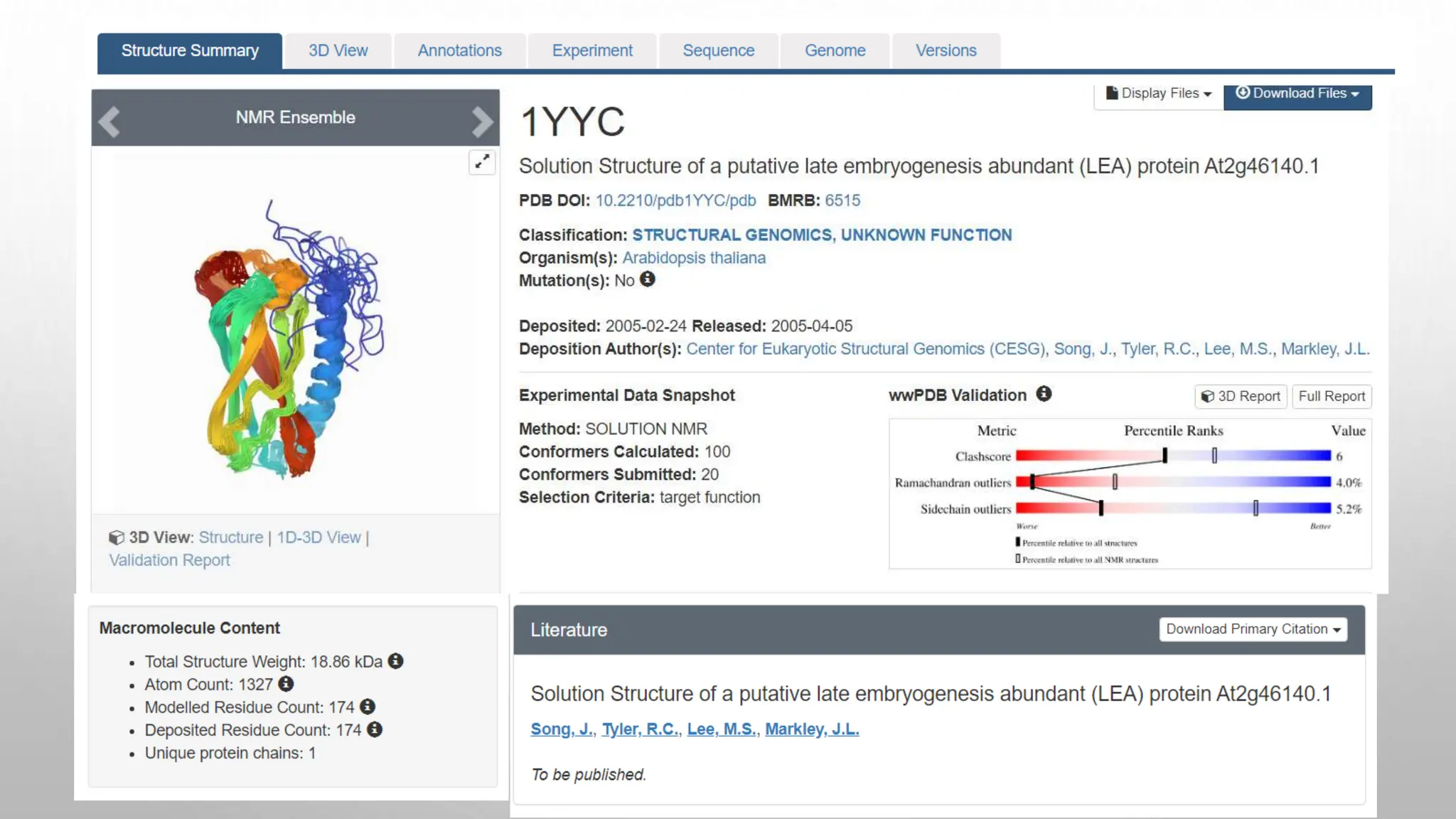
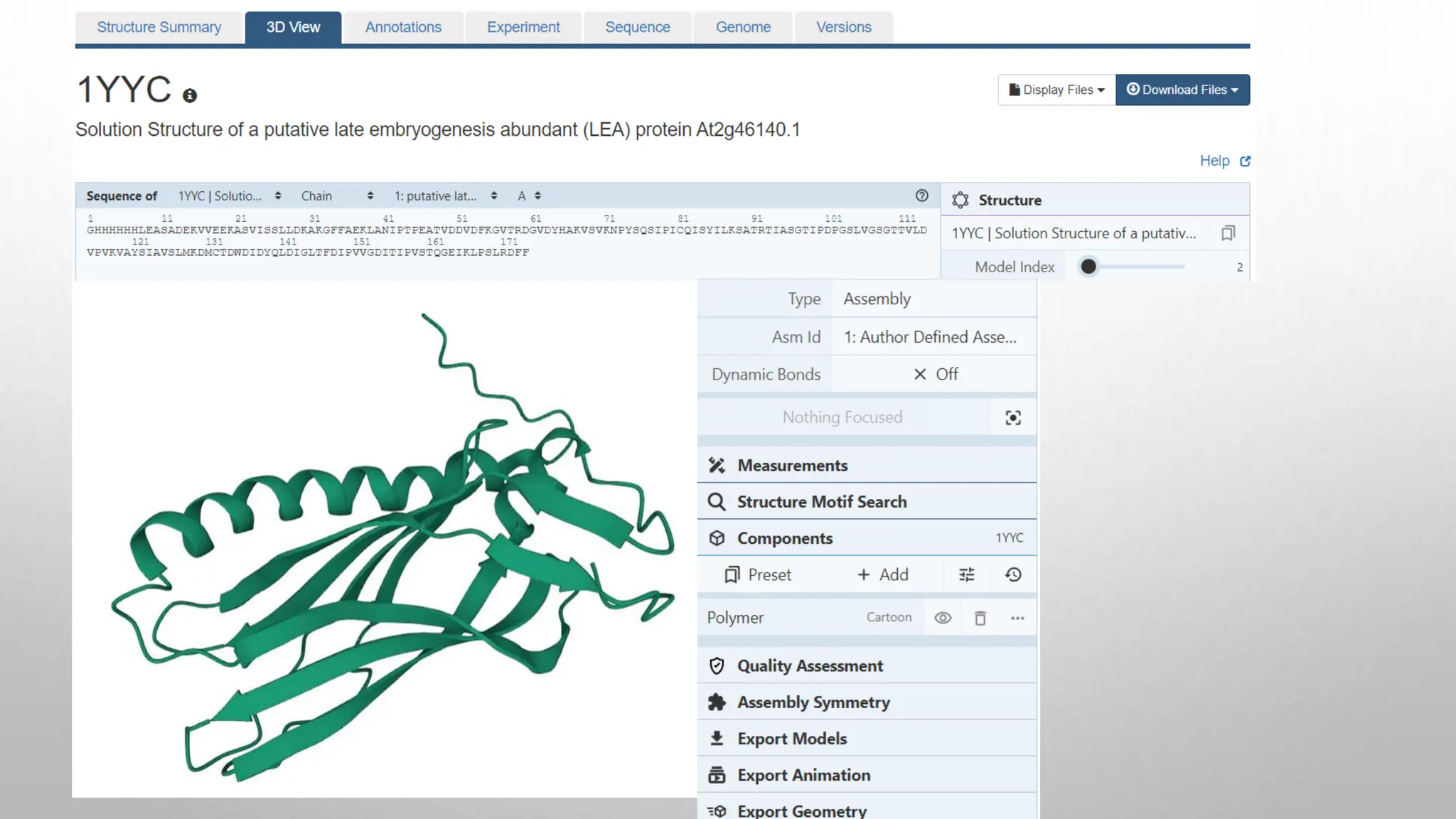
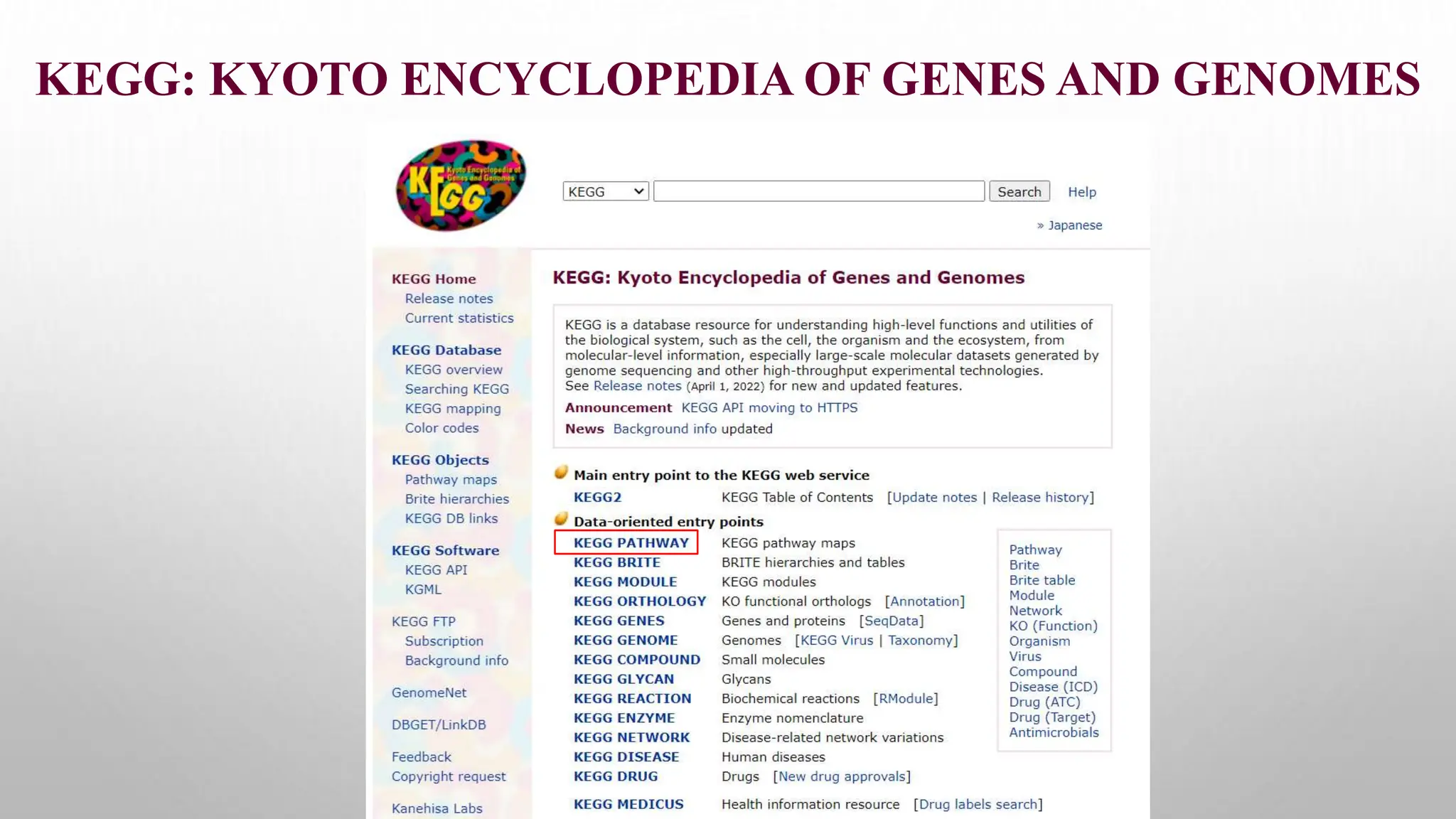
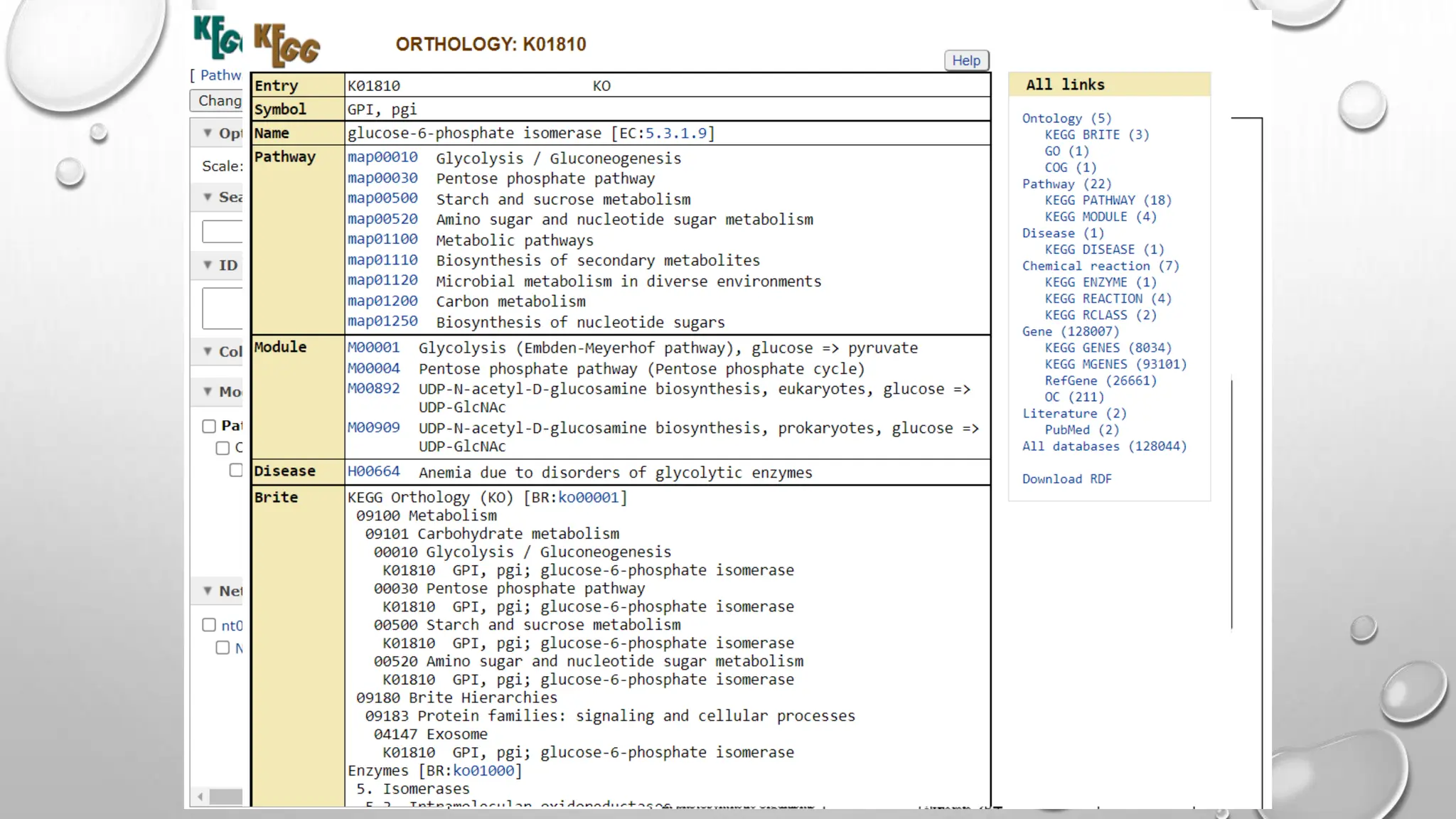
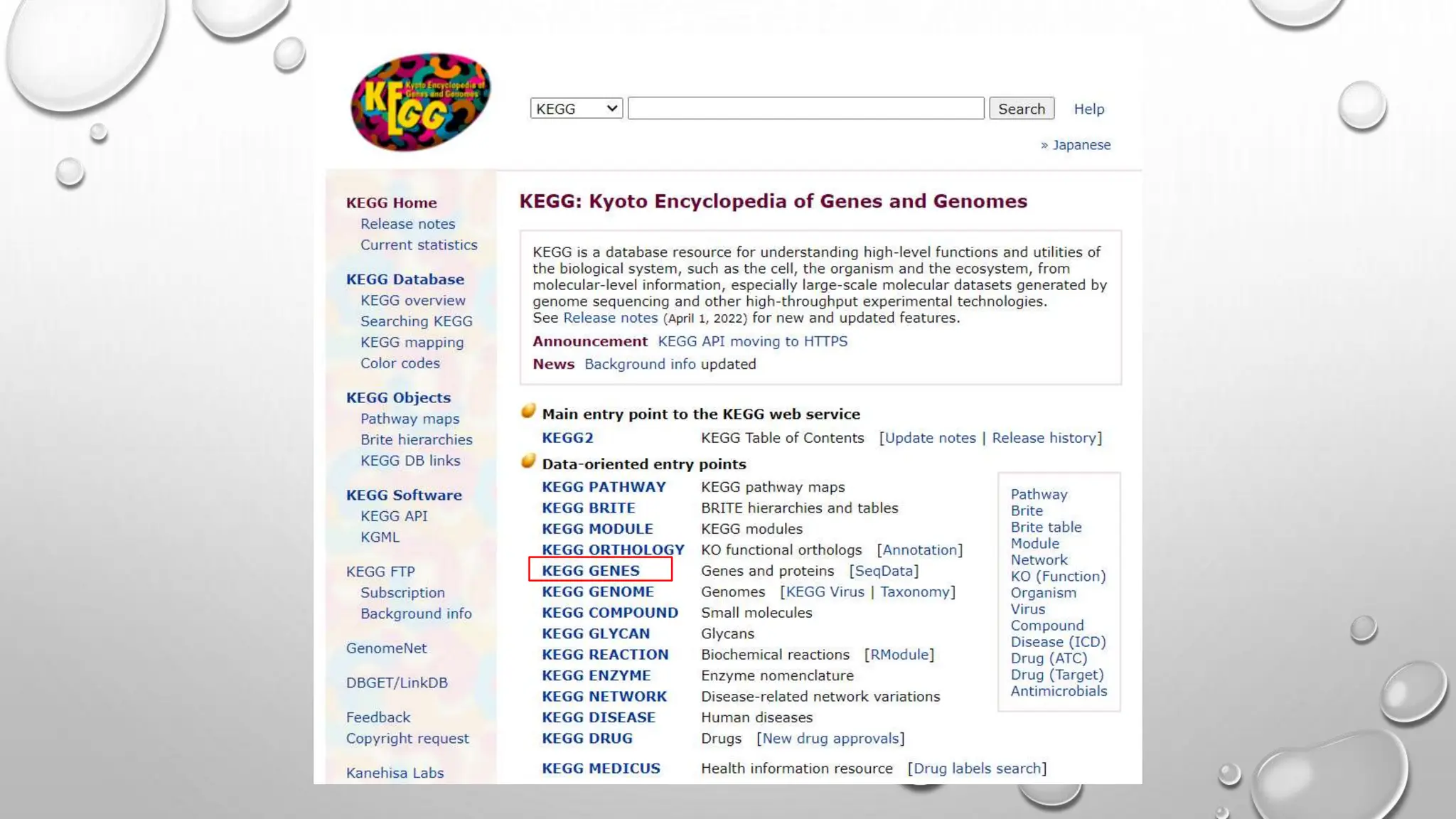
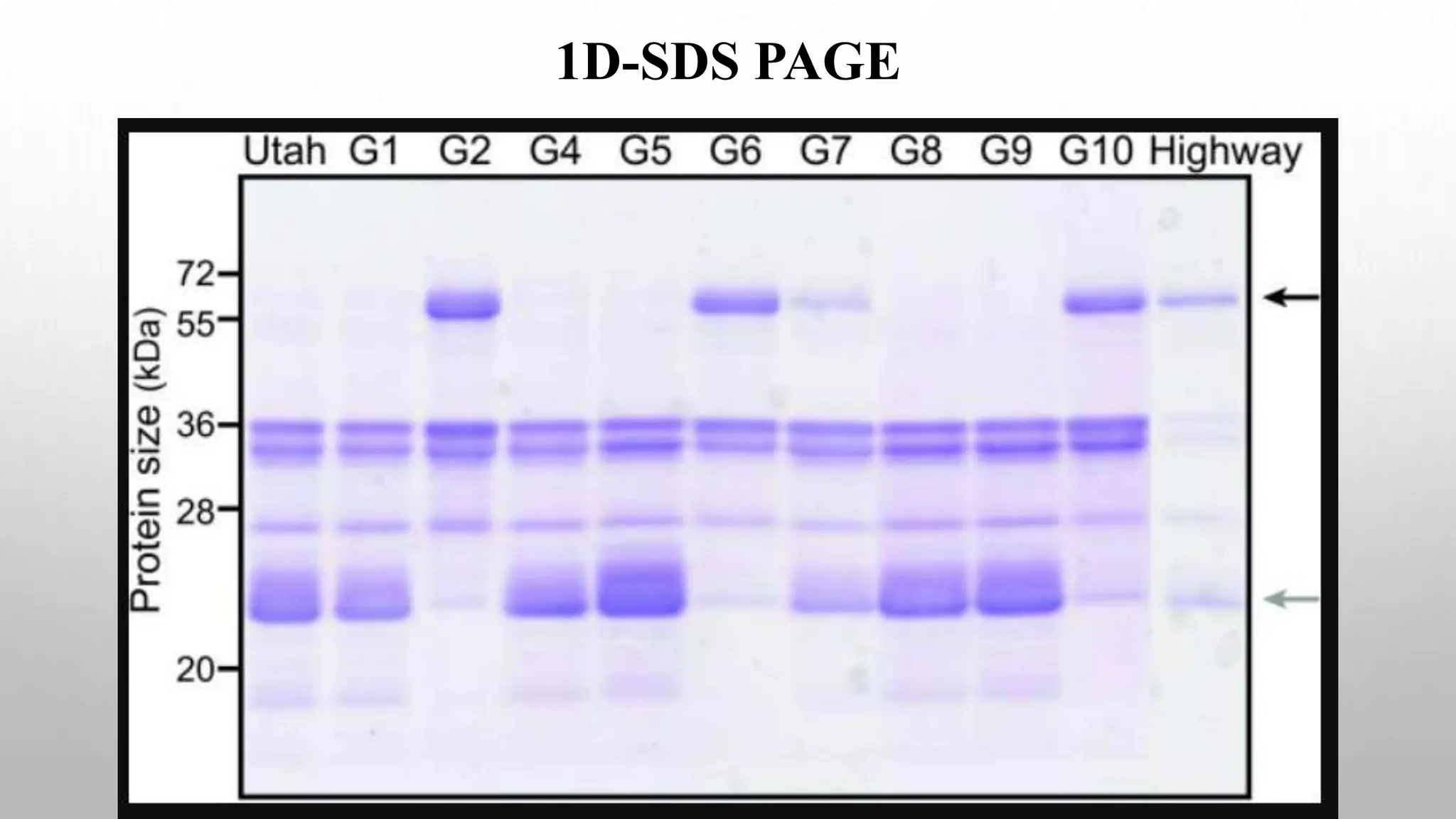
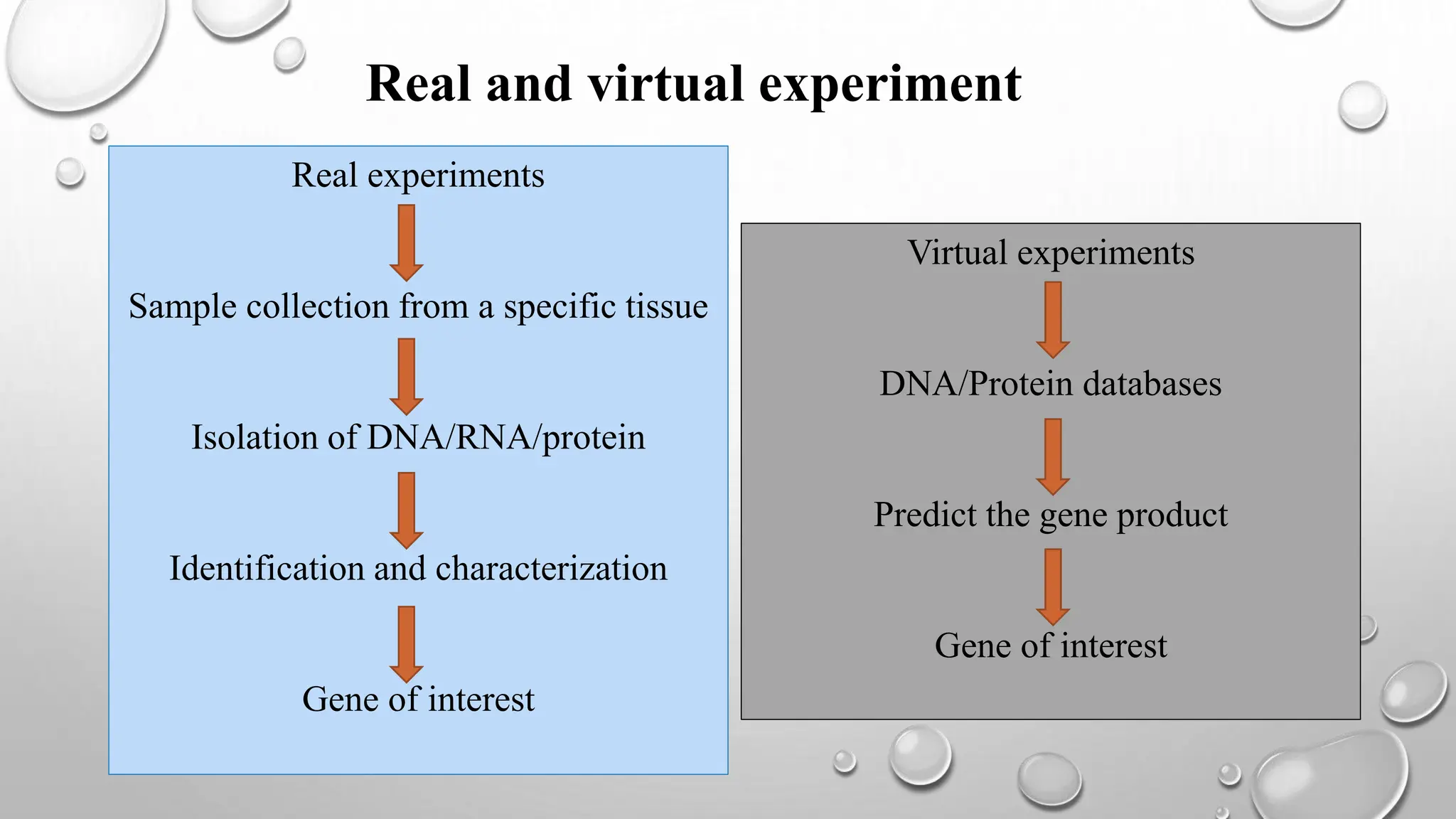
Proteomics techniques such as gel electrophoresis and mass spectrometry are used to separate and identify proteins. Two-dimensional gel electrophoresis separates proteins by size and charge, while MALDI-TOF mass spectrometry relies on mass spectrometry to analyze proteins and peptides. Protein-protein interactions can be studied using techniques like yeast two-hybrid systems and co-immunoprecipitation. Databases such as UniProt, PDB, and KEGG provide information on protein sequences, structures, and pathways.