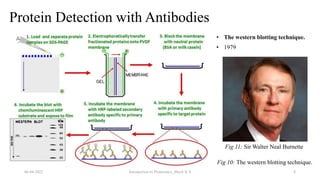
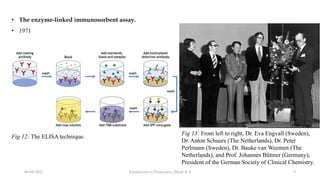
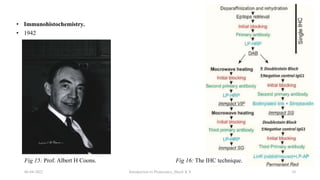
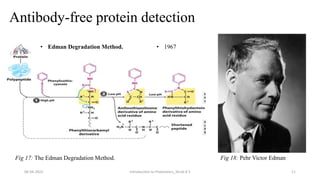
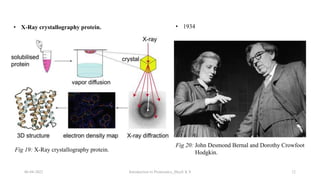
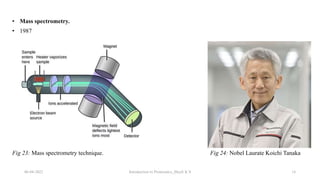
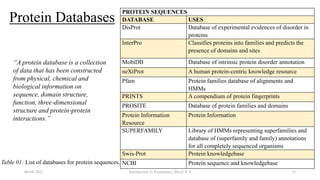
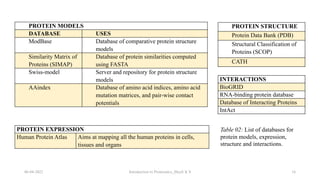
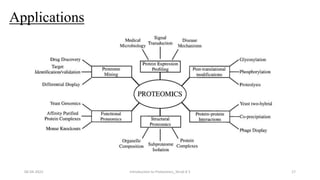
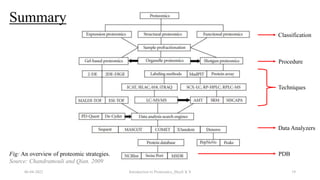
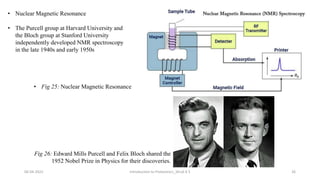
This document provides an introduction to proteomics presented by Shryli K S. It discusses the history of proteomics including early work using 2D gels in 1975. It describes the objectives of proteomics such as studying protein expression, function and interactions. Techniques used to study proteins are discussed including protein detection using antibodies, mass spectrometry, and protein databases. The conclusion recognizes important Nobel Prize winners that advanced the field of proteomics and proteomic applications.