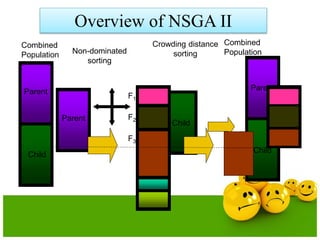
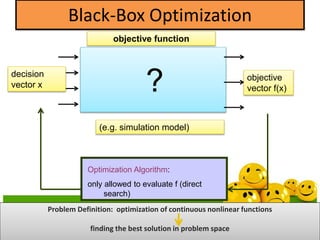
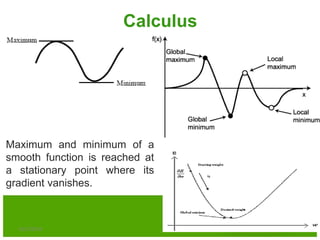
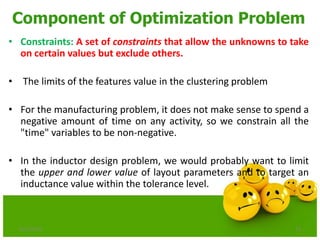
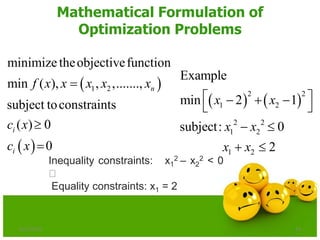
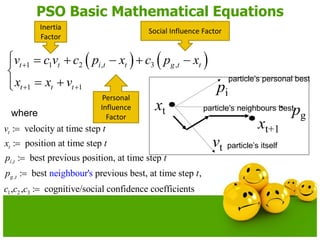
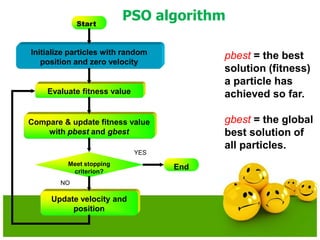
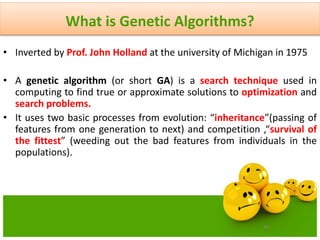
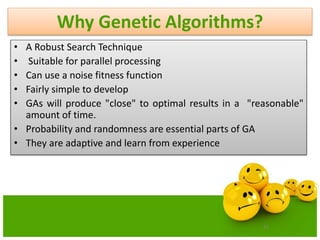
The document discusses various optimization techniques including evolutionary computing techniques such as particle swarm optimization and genetic algorithms. It provides an overview of the goal of optimization problems and discusses black-box optimization approaches. Evolutionary algorithms and swarm intelligence techniques that are inspired by nature are also introduced. The document then focuses on particle swarm optimization, providing details on the concepts, mathematical equations, components and steps involved in PSO. It also discusses genetic algorithms at a high level.






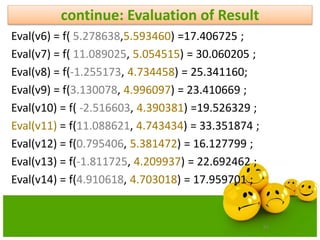
![20
PSO Velocity Update Equations Using
Constriction Factor Method
0.729)Kso4.1,set towas(
4,
42
2
)]()([
21
2
2211
cc
K
vxx
xprandcxprandcvKv
new
id
old
id
new
id
idgdidid
old
id
new
id](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-20-320.jpg)



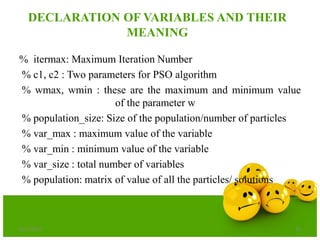

![Step-1: INITIALIZATION OF VARIABLES
itermax = 100;
c1 = 2;
c2 = 2;
wmax = 0.9;
wmin = 0.4;
population_size = 20;
var_max = [5.12 5.12];
var_min = [-5.12 -5.12];
velocity_max = var_max;
velocity_min = var_min;
var_size =
length(var_max);
6/21/2013 28](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-28-320.jpg)

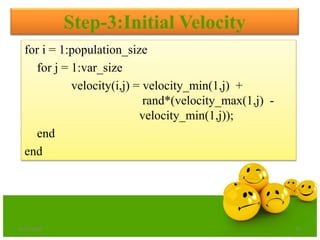
![fitness = objective_function(population);
pbest = population;
pbest_value = fitness;
[ xx yy] = min(fitness);
gbest = population(yy,:);
gbest_value = xx ;
Step-4:Determination of Pbest & Gbest
6/21/2013 31](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-31-320.jpg)

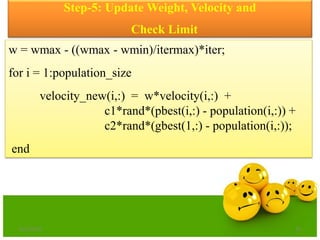


![Step-7: Modifying Pbest
fitness_new = objective_function(population_new);
[ x y] = min(fitness_new);
for i = 1:population_size
if fitness_new(i)< pbest_value
pbest(i,:) = population_new(i,:);
pbest_value(i) = fitness_new(i);
end
end
6/21/2013 36](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-36-320.jpg)


![Objective Function
function fitness = objective_function(population)
[row_population col_population] = size(population);
for i = 1: row_population
for j = 1: col_population
xx(j) = population(i,j);
end
fitness(i) = 20 + xx(1)^2 + xx(2)^2 -
10*(cos(2*pi*xx(1))+cos(2*pi*xx(2)));
end
6/21/2013 39](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-39-320.jpg)

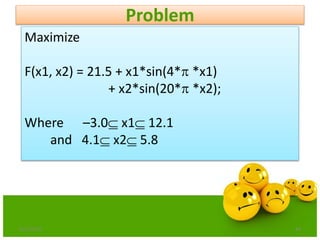
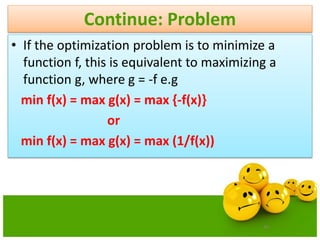
![Step:1 Representation
6/21/2013 46
Assume the required precision for each variable is upto
four decimal places.
The variable x1 has length 15.1 e.g [12.1 –3.0]
The precision requirement implies that the range[-3.0,
12.1] should be divided into at least 15.1*10000 equal size
ranges.
This means that 18 bits are required for the first part of the
chromosome.
217< 151000 <218](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-46-320.jpg)
![47
Continue: Representation
• The domain of variable x2 has length 1.7 e.g.[5.8-4.1]
• The precision requirement implies that the range [4.1,
5.8] should be divided into at least 1.7*10000 equal size
ranges.
• This means that 15 bits are required as the second part
of the chromosome.
214< 17000 <215](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-47-320.jpg)







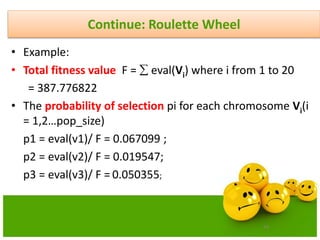
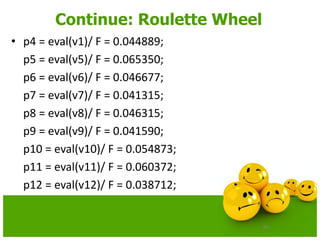
![58
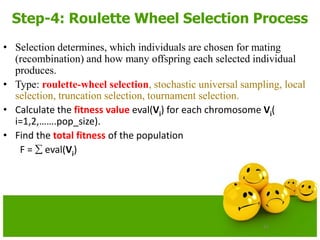
Continue: Roulette Wheel
• Calculate the probability of a selection pi for each chromosome Vi
(I,2,……pop_size).
pi = eval(Vi)/ F
• Calculate the cumulative probability qi
for each chromosome Vi(I = 1,2,… pop_size);
qi = pj where j varies from 1 to i
• Generate a random (float) number r from the range [0..1]
• If r<q1then select the first chromosome(v1); otherwise select the ith
chromosome Vi (2 i pop_size) such that q i-1 < r qi.](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-58-320.jpg)

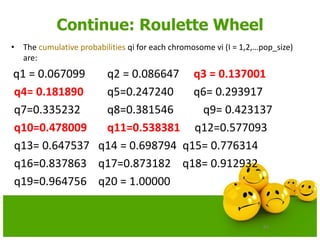
![63
Continue: Roulette Wheel
• Let us assume that a (random) sequence of 20 numbers from the
range[0..1]is:
0.513870 0.175741 0.308652 0.534534 0.947628
0.171736 0.702231 0.226431 0.494773 0.424720
0.703899 0.389647 0.277226 0.368071 0.983437
0.005398 0.765682 0.646473 0.767139 0.780237](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-63-320.jpg)




![68
Continue: Crossover
• Assume the probability of crossover Pc.
• This probability gives us the expected number Pc*
pop_size of chromosomes which undergo the
crossover operation.
• Generate a random(float) number r from the
range[0..1]
• If r<Pc, select the given chromosome for
crossover](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-68-320.jpg)






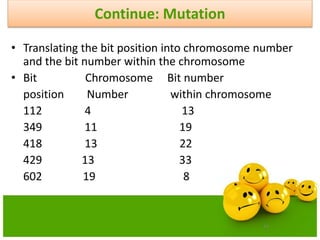
![75
Continue: Mutation
• Mutation is performed bit-by-bit basis
• Assume the probability of Mutation Pm.
• This probability gives us the expected number of
mutated bits Pm* pop_size.
• Generate a random(float) number r from the
range[0..1]
• If r<Pm, mutate the bit](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-75-320.jpg)











![6/21/2013 90
]10,0[
2010
;)10()(
;)(
2
2
2
1
x
utionOptimalSol
x
where
xxf
xxf
Minimize](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-90-320.jpg)
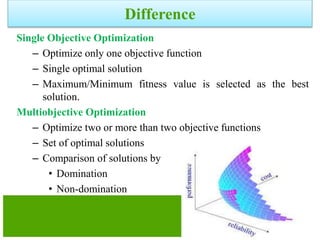
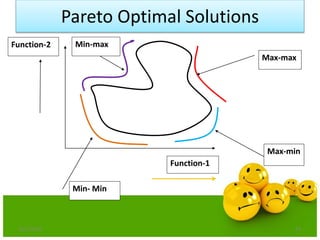
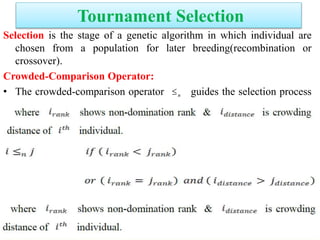
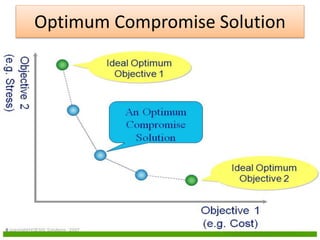
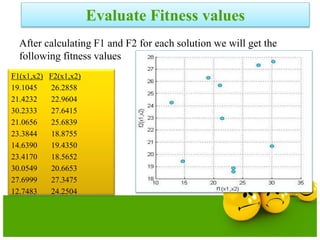
![Definitions
Domination: One solution is said to
dominate another if it is better in all
objectives.
Non-Domination[Pareto points]: A solution
is said to be non-dominated if it is better
than other solutions in at least one
objective
•A dominates B (better in both ƒ1 and ƒ2)
•A dominates C (same in ƒ2 but better in ƒ1)
•A does not dominate D (non-dominated points)
•A and D are in the “Pareto optimal front”
•These non-dominated solutions are called Pareto optimal solutions.
•This non-dominated curve is said to be Pareto front.](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-95-320.jpg)
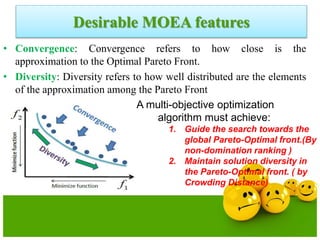

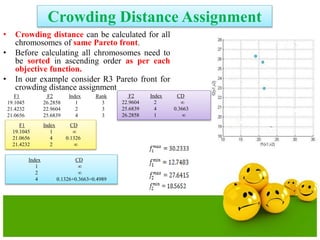
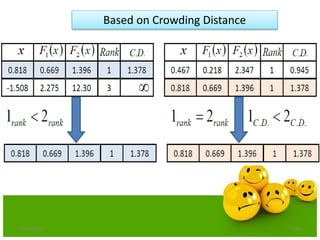
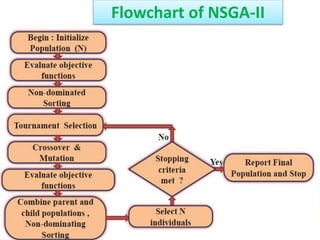
![Crowding Distance Assignment
• To get an estimate of density of
solutions surrounding a particular
solution in population.
• Choose individuals having large
crowding distance.
• Help for obtaining uniformly
distribution
where ƒ[i]m represent objective function value of solution. and are
the maximum and minimum value of the objective function.](https://image.slidesharecdn.com/cvraman-9-6-2015-final-200125082641/85/Optimization-Using-Evolutionary-Computing-Techniques-103-320.jpg)