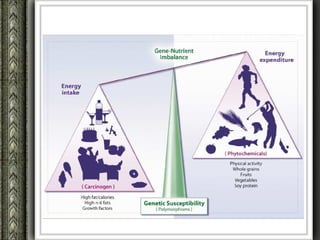
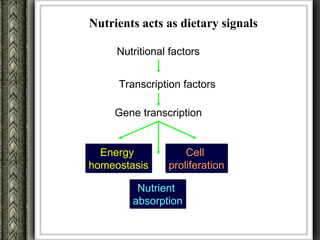
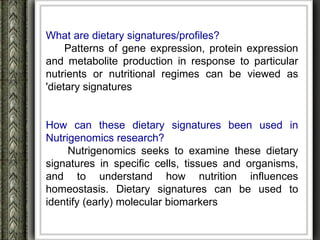
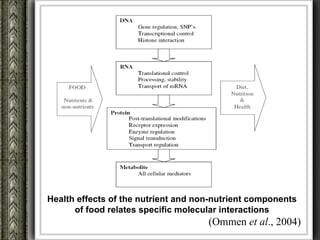
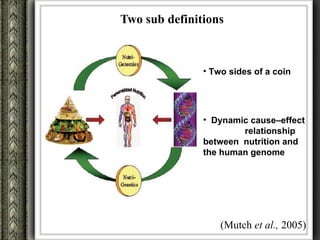
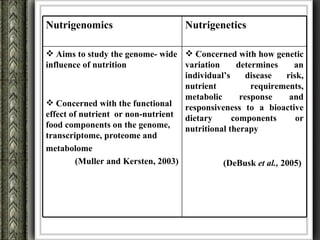
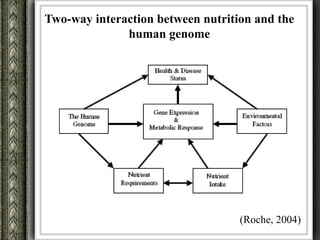
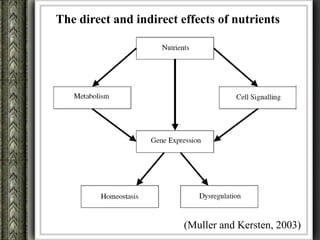
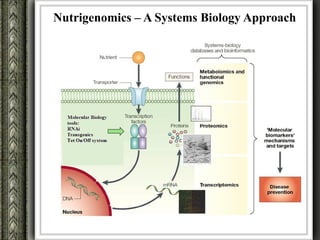
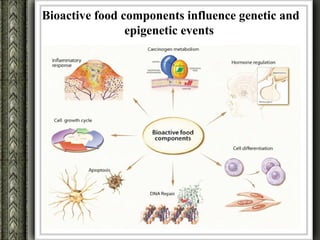
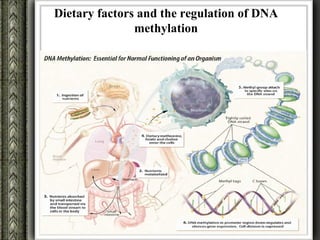
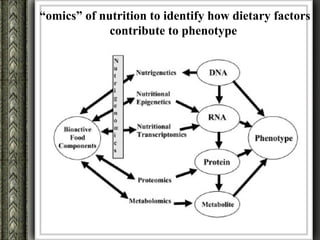
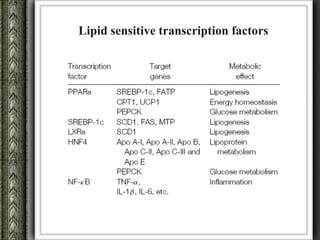
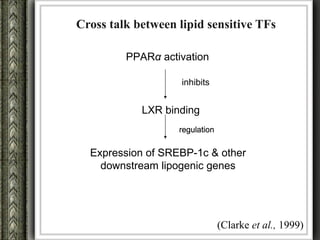
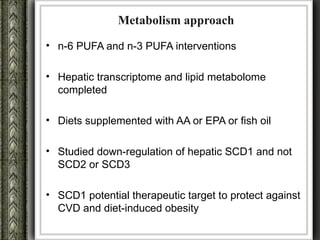
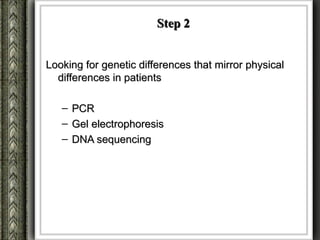
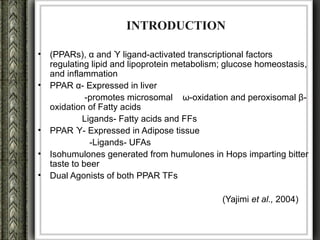
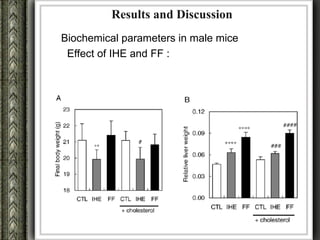
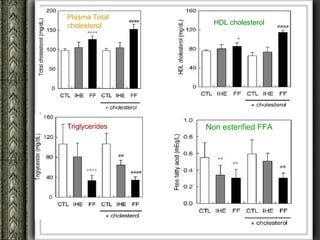
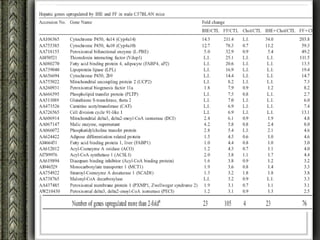
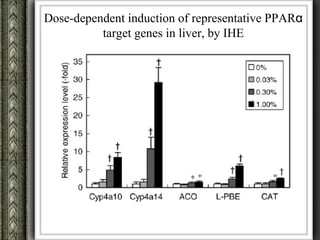
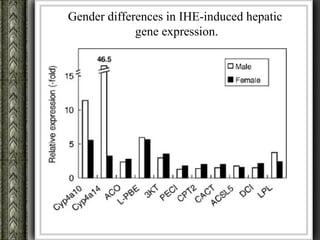
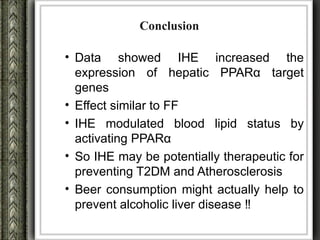
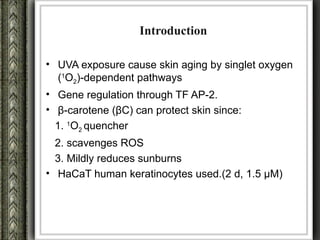
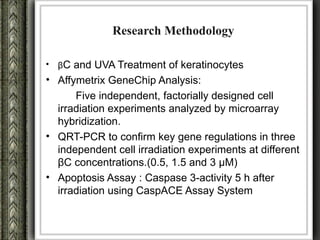
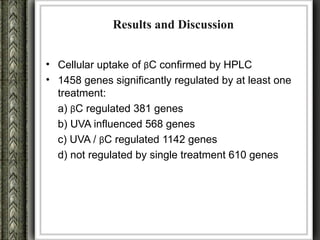
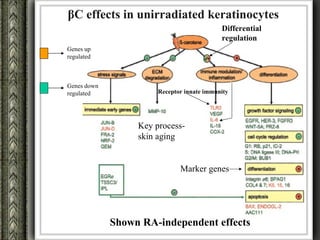
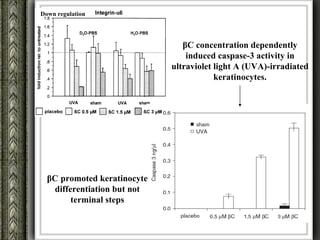
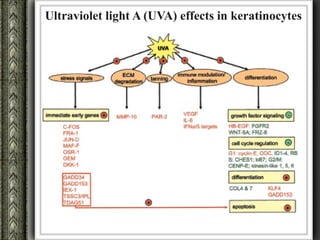
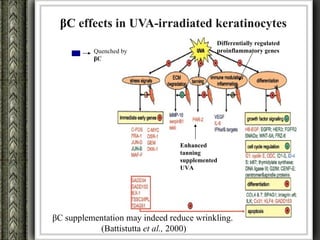
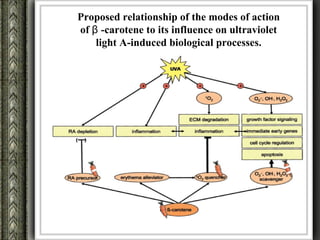
The document discusses nutrigenomics and nutrigenetics, emphasizing the dynamic relationship between nutrition and the human genome, including the role of dietary signatures in health and disease risk. It highlights the need for personalized nutrition approaches based on individual genetic variability and the implications for developing targeted dietary interventions. Additionally, it explores methodologies and findings related to the effects of specific nutrients on gene expression and metabolic responses.