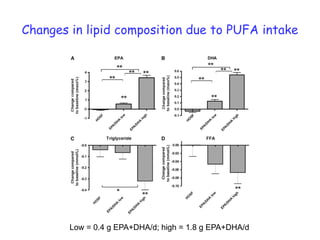
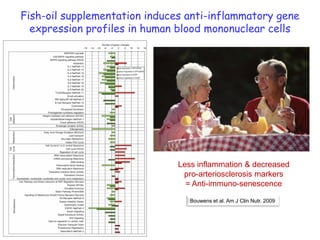
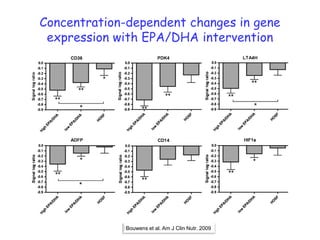
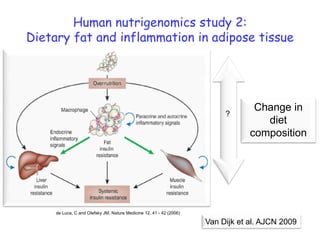
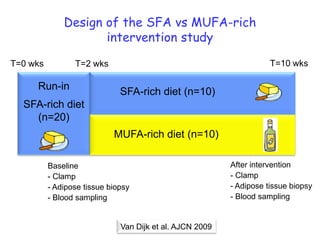
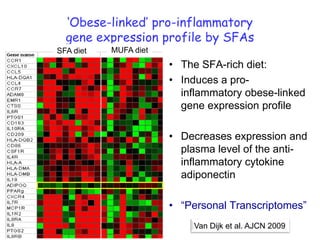
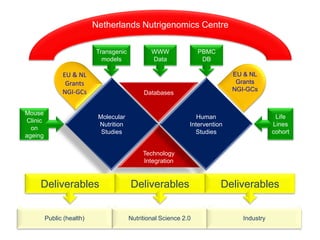
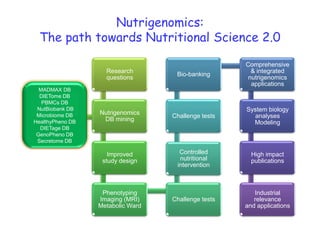
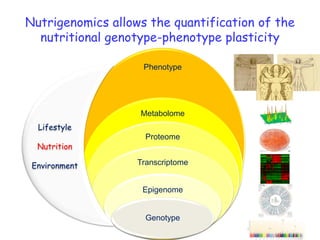
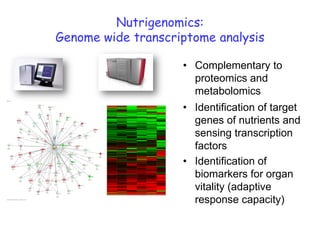
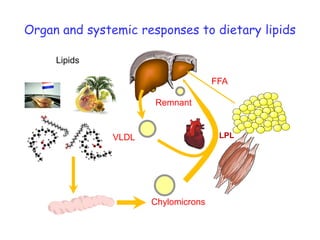
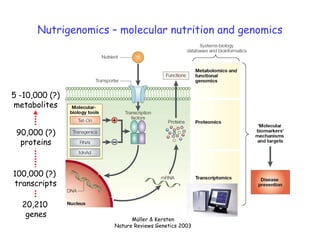
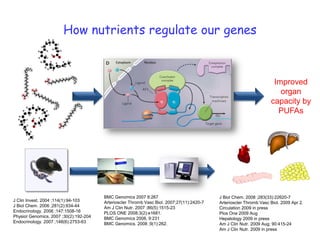
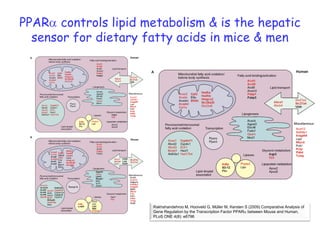
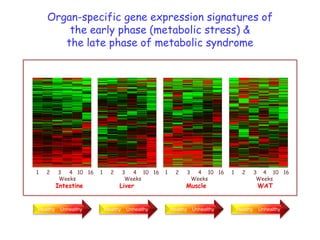
This document summarizes a lecture on nutrigenomics given by Michael Müller. It discusses how nutrigenomics allows quantification of the interplay between genetics, lifestyle, nutrition, and the environment in determining phenotypes. Specifically, it highlights how genome-wide transcriptome analysis can identify target genes of nutrients and biomarkers of organ health and resilience. It also provides examples of human nutrigenomics studies examining how diets high in saturated fat or polyunsaturated fat alter gene expression and inflammation. The summary concludes that nutrigenomics is enabling a transition to nutritional science 2.0 through comprehensive, integrated applications and system biology analyses.





![You are what you (not) eat
24h 48h
PATHWAYS up down up down
carnitine O-palmitoyltransferase activity
O-palmitoyltransferase activity
palmitoyltransferase activity
3-oxoacyl-[acyl-carrier protein] reductase activity
Fatty acid metabolism fatty-acid synthase activity
carnitine O-acyltransferase activity
fatty acid beta-oxidation
fatty acid oxidation
acetyl-CoA C-acyltransferase activity
lipoate biosynthesis
Pyruvate metabolism pyruvate transporter activity
heterogeneous nuclear ribonucleoprotein complex
regulation of viral genome replication
RNA processing
RNA/DNA metabolism B-cell differentiation
negative regulation of transcription
small ribosomal subunit
cytoplasmic microtubule
glyoxylate metabolism
heme biosynthesis
Other histone-lysine N-methyltransferase activity
procollagen N-endopeptidase activity
alpha-ketoglutarate dehydrogenase complex
TCA cycle TCA cycle enzyme complex (sensu Eukarya)
up_24h down_24h neg up_48h down_48h neg
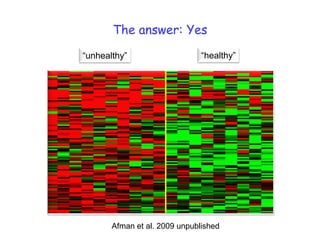
Bouwens et al. Am J Clin Nutr. 2007](https://image.slidesharecdn.com/icncascadelecturemullerfinal-100108020443-phpapp02/85/Nutrigenomics-The-path-towards-nutritional-science-2-0-16-320.jpg)