The document discusses the genetic and molecular mechanisms underlying the flowering process in plants, emphasizing the roles of floral meristem identity and floral organ identity genes in determining flower structure. It describes the classical ABC model of floral organ identity, where the interaction of classes of genes specifies the development of sepals, petals, stamens, and carpels. The document also highlights the involvement of microRNAs and MADS-box genes in regulating floral development and transitions between vegetative and reproductive phases.

![Genes That Regulate Floral Development
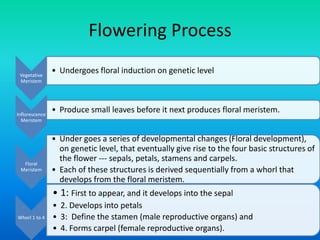
• Two major categories :
– Floral meristem identity genes : Activity of these
genes, in the immature primordia formed at the
flanks of the shoot apical meristem (inflorescence
meristem), results in its transition to form the
floral meristem.
– Floral organ identity genes: Floral organ identity
genes directly control floral organ identity.
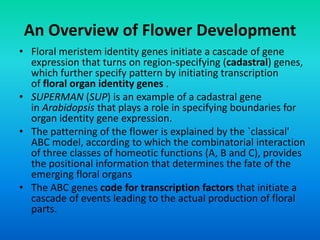
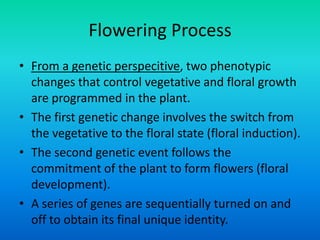
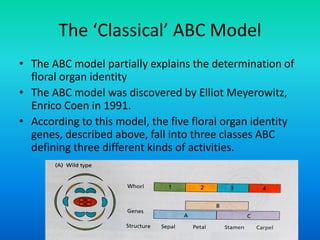
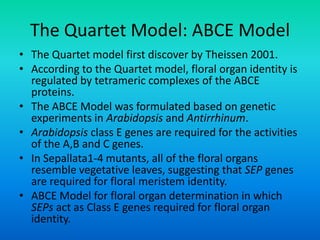
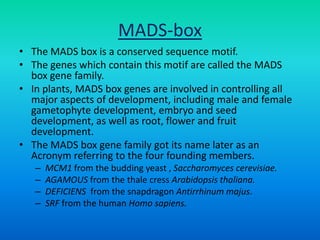
• {Five key genes initially were identified in Arabidopsis
that specify floral organ identity. APETALA1 [AP1],
APETALA2 [AP2], APETALA3 [AP3], PISTILLATA [PI] and
AGAMOUS [AG].}](https://image.slidesharecdn.com/floraldevelopment-200510210647/85/Floral-development-5-320.jpg)
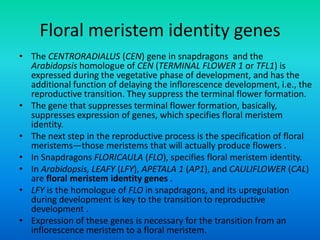
![Floral organ identity genes
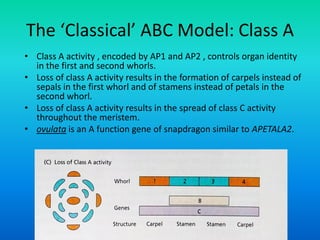
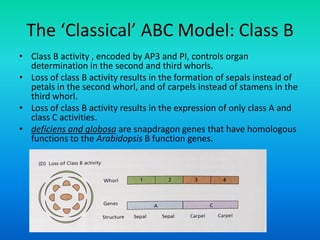
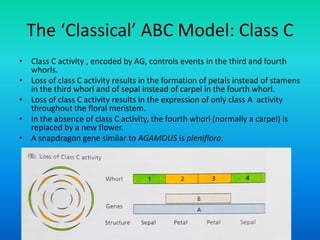
• Floral organ identity genes
– Floral organ identity genes directly control floral organ
identity.
– Five key genes initially were identified in Arabidopsis that
specify floral organ identity. APETALA1 [AP1], APETALA2
[AP2], APETALA3 [AP3], PISTILLATA [PI] and AGAMOUS
[AG].
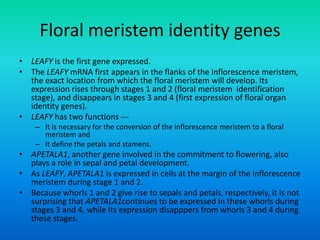
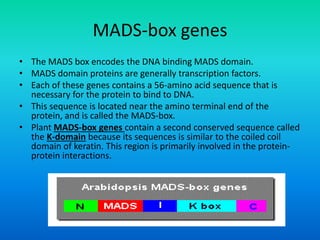
• These genes are homeotic genes.
• All homeotic genes that have been identified so far, in
plants, encode transcription factors.
• Most plant homeotic genes belong to a class of related
sequences known as MADS box genes.](https://image.slidesharecdn.com/floraldevelopment-200510210647/85/Floral-development-8-320.jpg)
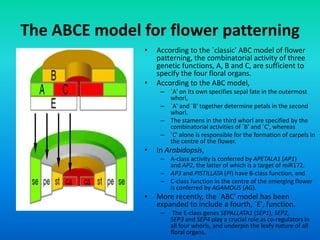
![Class D Genes Are Required For Ovule Formation.
• Class D activities were first discovered in petunia.
• Silencing, MADS box genes known to be involved in floral
development in petunia, FLORAL- BINDING
PROTEIN7/11[FBP7/11] , resulted in the growth of styles
and stigmas in the locations normally occupied by ovules.](https://image.slidesharecdn.com/floraldevelopment-200510210647/85/Floral-development-15-320.jpg)
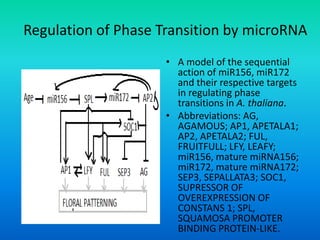


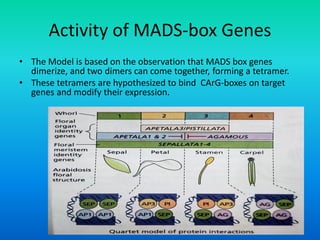
![Activity of MADS-box Genes
• The MADS box gene transcription factors form tetramers that
bind to CC[A/T]6GG sequences, the so called CArG-box, in the
regulatory regions of their target genes.
• When the tetramers bind two different CArG-boxes on the same
target gene, the boxes are brought into close proximity, causing
DNA bending.](https://image.slidesharecdn.com/floraldevelopment-200510210647/85/Floral-development-22-320.jpg)
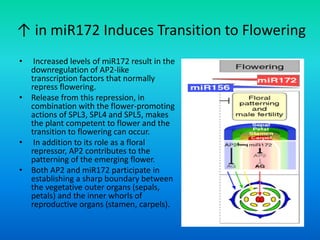
![Activity of MADS-box Genes
• Although all MADS box proteins can form higher order
complexes, not all of these are able to bind DNA.
• For example, Class B factors [AP3 and PI] bind DNA only
as heterodimers, whereas both homodimers and heterodimers
of Classes A, C and E can bind DNA.
• According to the Model, tetramers composed of different
homodimers and heterodimers of MADS domain proteins
can exert combinatorial control over floral organ identity.](https://image.slidesharecdn.com/floraldevelopment-200510210647/85/Floral-development-24-320.jpg)