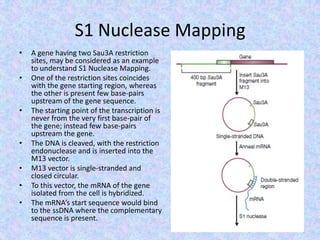
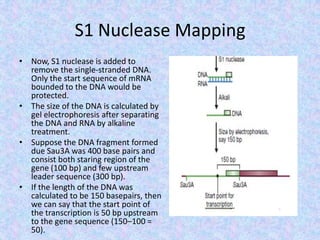
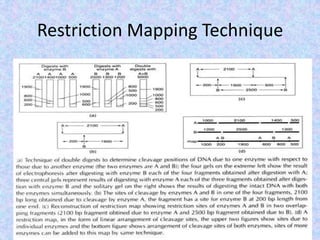
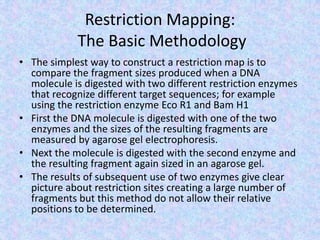
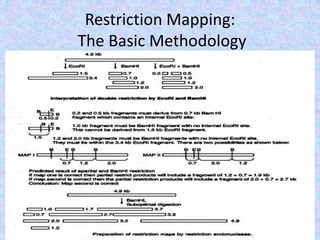
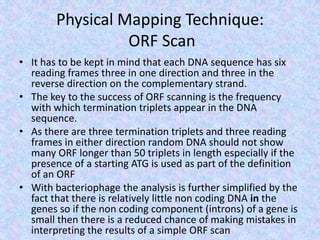
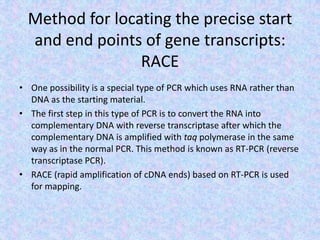
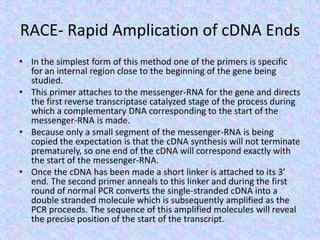
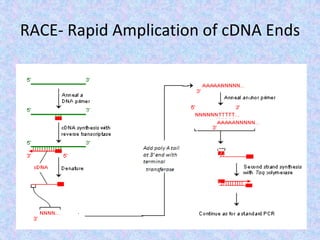
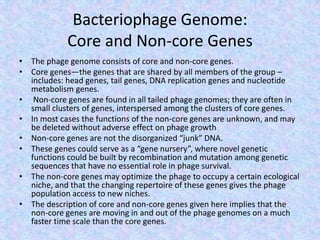
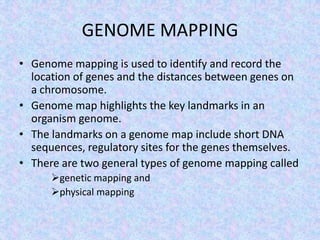
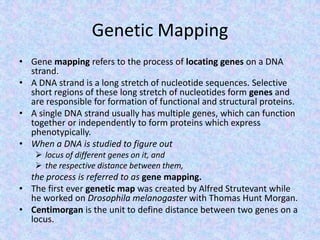
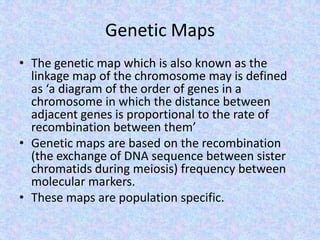
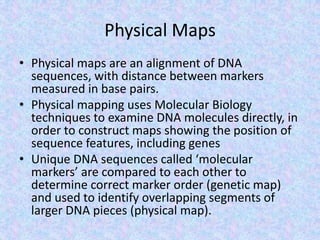
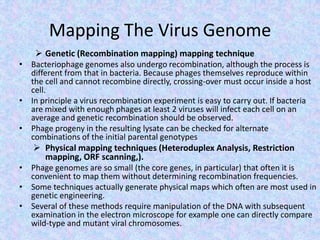
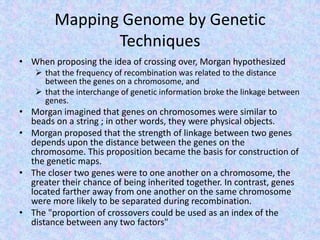
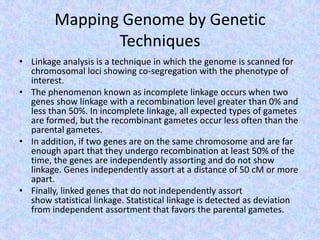
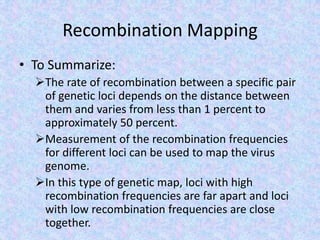
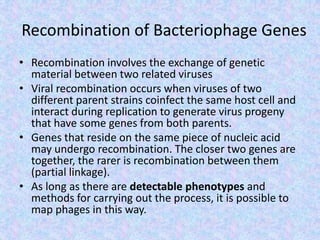
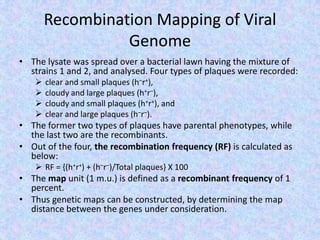
The document discusses the diverse and mosaic nature of bacteriophage genomes, detailing their composition of core and non-core genes and the importance of genome mapping for identifying gene locations. It elaborates on genetic and physical mapping techniques used for understanding recombination and constructing maps of viral genomes, including methodologies like heteroduplex analysis and restriction mapping. Additionally, it highlights the implications of these mapping techniques for understanding viral evolution and gene function.



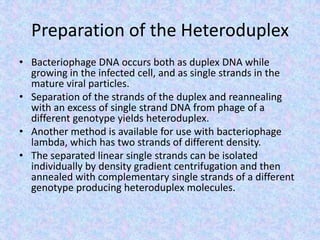
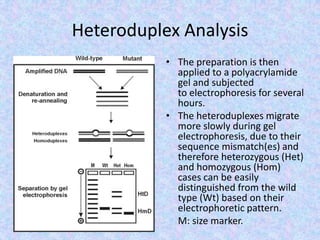
![Heteroduplex Analysis
• Two different methods are applied under the
concept of heteroduplex analysis.
• With the molecular biological method, a standard
DNA and a DNA to be analyzed are amplified
separately, mixed, denatured, and slowly cooled
(approximately 1°C to 2°C [34°F to 37°F] per
minute) to permit the formation of both,
homoduplexes (HmD) and heteroduplexes (HtD).
Heteroduplexes are formed from standard and
sample DNA.](https://image.slidesharecdn.com/mappingthebacteriophagegenome-200502164710/85/Mapping-the-bacteriophage-genome-18-320.jpg)