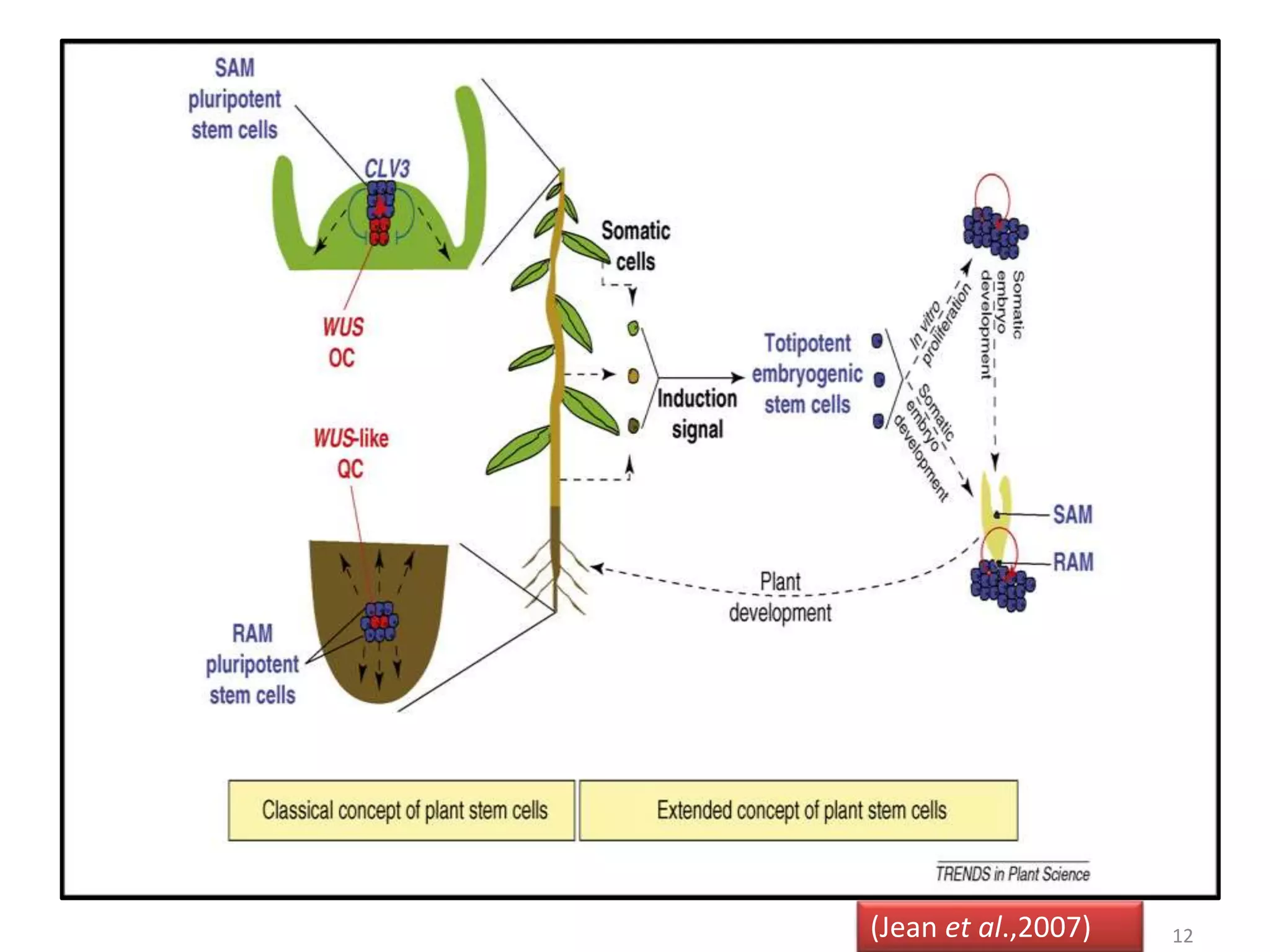
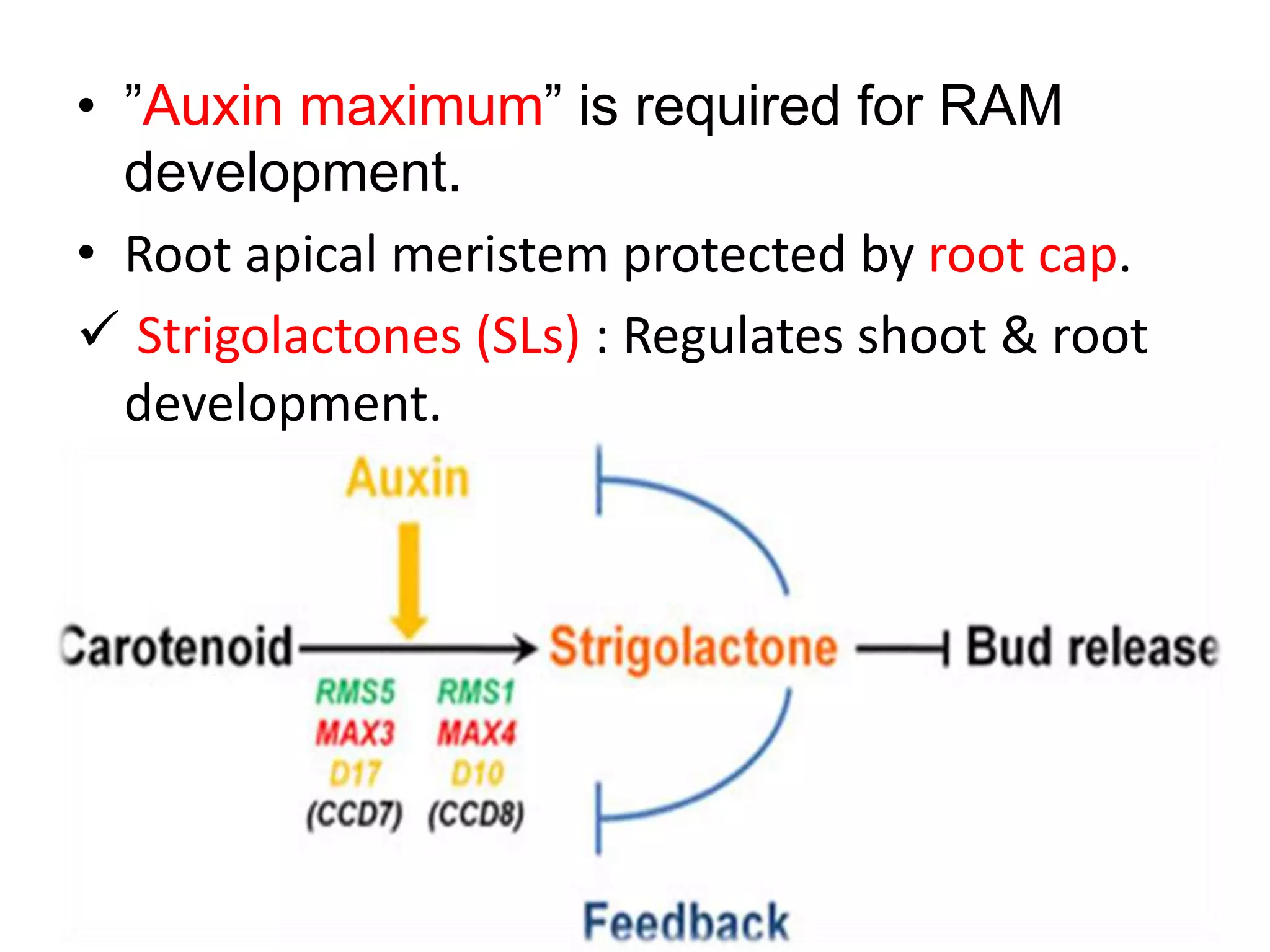
The document discusses meristematic tissues and apical meristems in plants. It summarizes that the shoot apical meristem (SAM) and root apical meristem (RAM) contain stem cells and are responsible for postembryonic growth. The SAM contains four distinct cell groups and is maintained by genes like SHOOT MERISTEMLESS, WUSCHEL, and CLAVATA1/3. The RAM contains a quiescent center and produces root cells. Key genes that regulate SAM and RAM development include MONOPTEROS and HOBBIT.