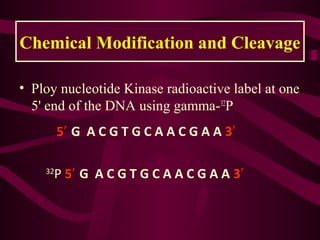
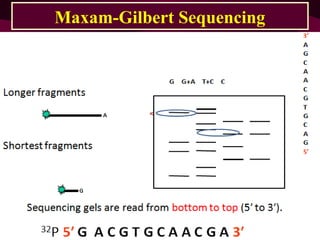
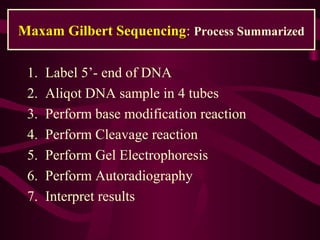
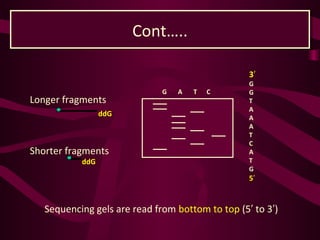
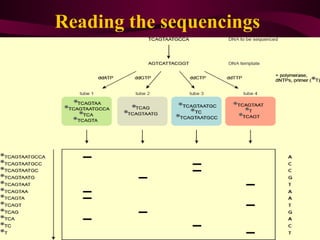
This document provides information about DNA sequencing methods. It discusses that DNA sequencing determines the precise order of nucleotides in DNA. The two main conventional methods described are the Maxam-Gilbert chemical degradation method and the Sanger chain termination method. The Maxam-Gilbert method uses chemical modification and cleavage of DNA bases. The Sanger method is a PCR-based method that uses dideoxynucleotides to terminate DNA chain elongation. Both methods produce fragmented DNA of different lengths that can be resolved on a gel for sequence determination.