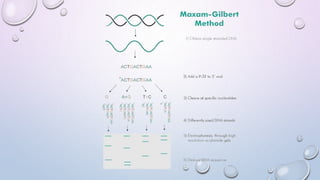
The document provides a comprehensive overview of gene and genome sequencing, detailing the importance of DNA sequencing methods such as Maxam–Gilbert and Sanger sequencing. It highlights the structure and function of DNA, the process of sequencing, and the evolution of sequencing technologies, including next-generation methods. The discussion includes the applications and limitations of each method, underscoring Sanger sequencing's significance in validating DNA sequences.