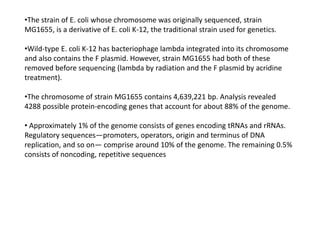
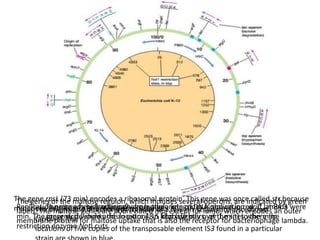
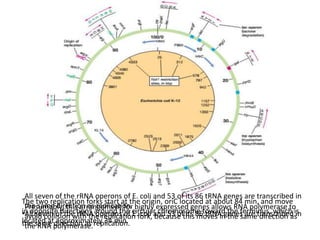
This document summarizes bacterial and viral chromosomes. It discusses the structure of bacterial chromosomes, using E. coli as an example. It notes that E. coli has a single circular chromosome containing all of its genes. It describes the sequencing and mapping of the E. coli chromosome, including its size, gene content, and organization. It also discusses horizontal gene transfer in E. coli and the insertion of genetic elements into its chromosome. The document then summarizes viral genomes, noting that viruses have either DNA or RNA and can be single or double stranded. It provides details on the genome of bacteriophage T4.