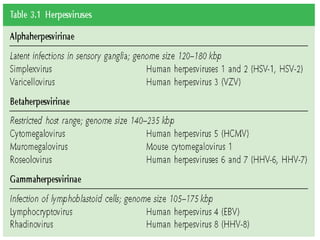
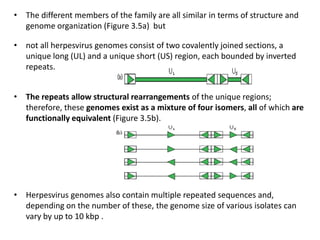
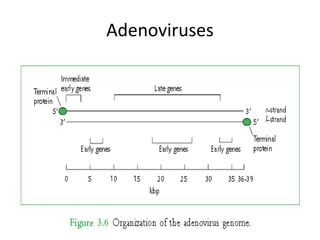
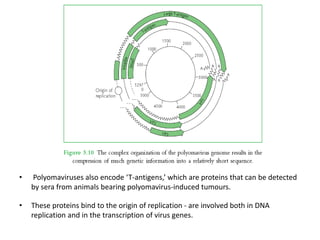
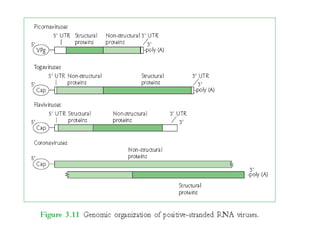
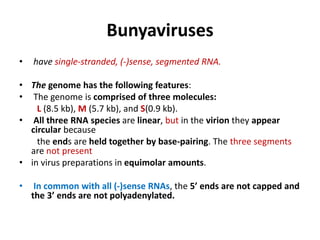
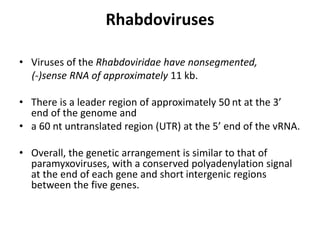
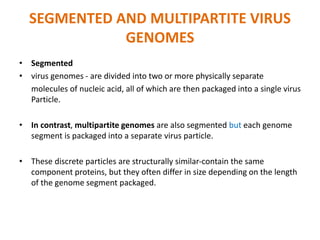
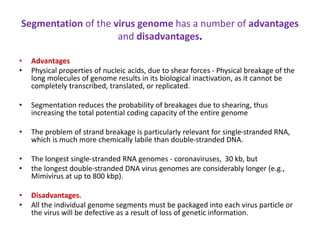
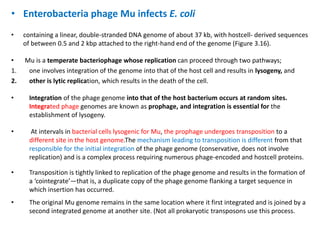
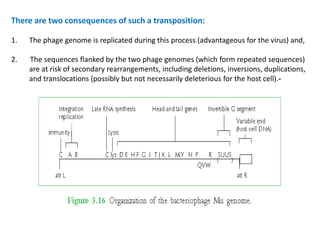
The document discusses viruses with large and small DNA genomes as well as positive-strand and negative-strand RNA viruses. Herpesviruses have very large DNA genomes up to 235 kbp that encode many enzymes. Adenoviruses and phages like lambda have smaller genomes between 30-54 kbp. Animal viruses like parvoviruses and polyomaviruses have even smaller genomes around 5 kbp that tightly pack genes. Picornaviruses, togaviruses, and flaviviruses have single-stranded RNA genomes between 7-11 kbp. Coronaviruses have the largest RNA genomes around 30 kbp. Segmentation allows larger coding capacity for viruses like influenza and gemin