This document discusses gene synthesis techniques and blotting methods. It provides details on:
1) The first chemical synthesis of genes in the 1970s, including a gene for yeast tRNA and bacterial tRNA.
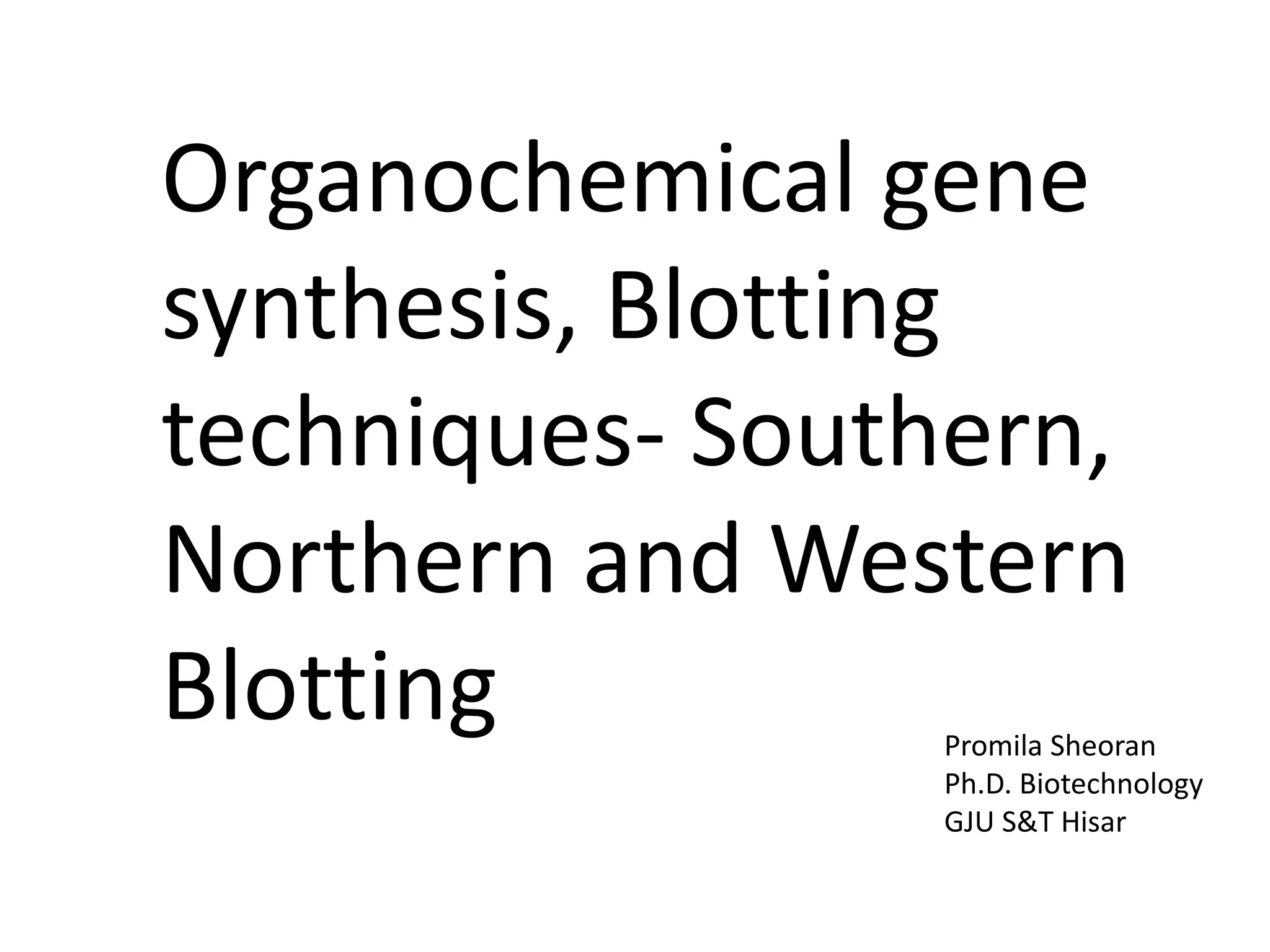
2) Methods for artificially synthesizing genes using oligonucleotides and ligating DNA fragments.
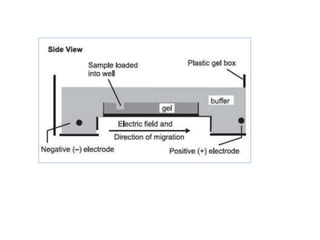
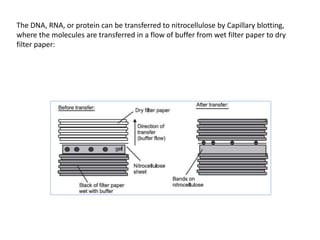
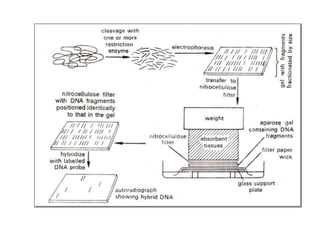
3) Techniques for analyzing DNA, RNA, and proteins - Southern blotting detects DNA, Northern blotting detects RNA, and Western blotting detects proteins.