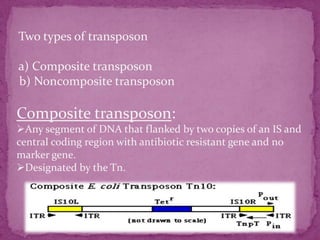
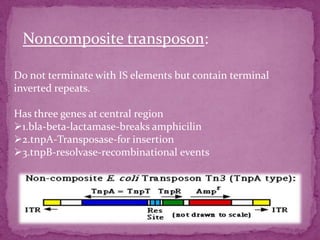
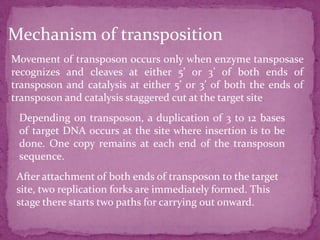
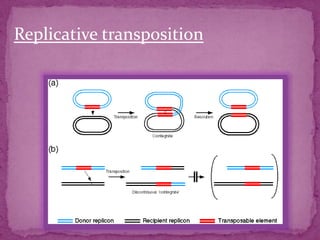
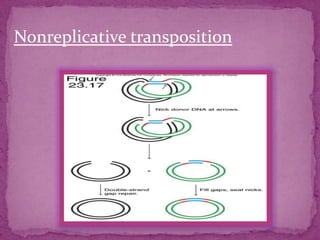
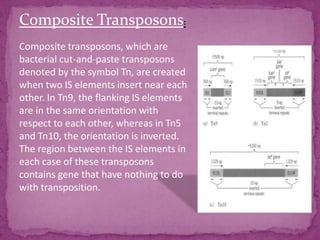
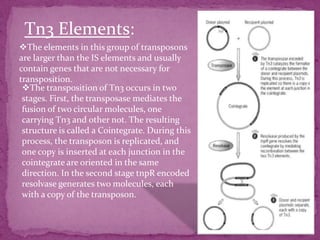
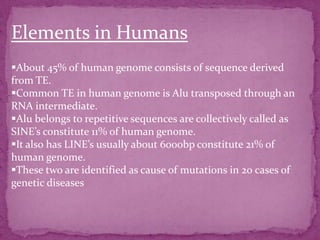
This document discusses transposable elements (TEs), which are segments of DNA that can move within genomes. It covers their discovery by Barbara McClintock in corn in the 1940s. TEs are classified into different types based on their structure and mechanism of movement. The document also examines the mechanisms of transposition, mutagenic effects, regulation, and presence of TEs across bacteria, fungi, and eukaryotes like humans. TEs make up a large fraction of genomes and contribute to genetic variation and disease.



![Nomenclature
Campbell et al in 1977 described the nomenclature in
prokaryotes.
Initially named as insertion sequences- IS IS1, IS2, IS3etc.,
In bacteria transposons containing genes for antibiotic
resistance are named as Tn like Tn1, Tn2, etc.
The number distinguish different transposons, those are
represented by standard genotypic designation such as Tn1
[ampr], where ampr refers that the transposons carries the
genetic locus or Ampicilin resistance
In eukaryotes named in nonstandard way
ex: Drosophila- copia,497, p-elements
yeast- Ty, Maize- Ds& Ac , Human-Alu](https://image.slidesharecdn.com/presentationseminar10-10-2011-120110080330-phpapp02/85/TRANSPOSABLE-ELEMENTS-5-320.jpg)