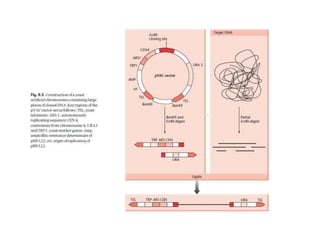
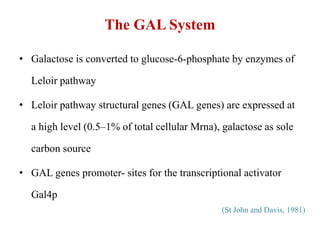
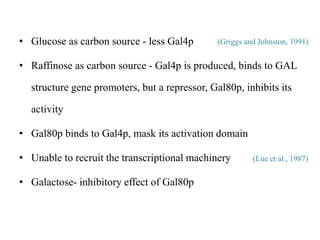
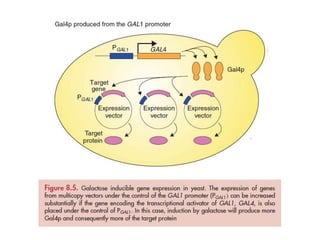
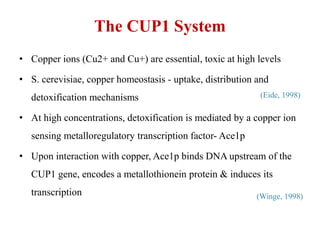
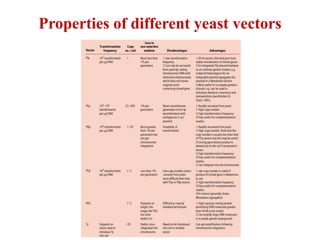
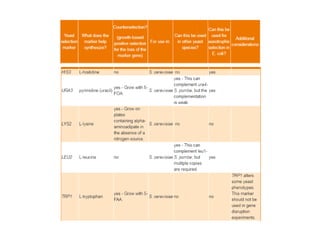
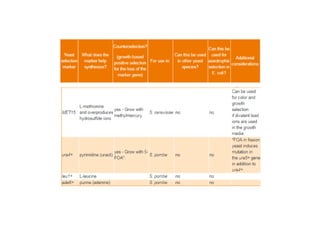
Yeast vectors are useful for expressing eukaryotic proteins due to yeasts' ability to perform post-translational modifications. Common yeast species used include Saccharomyces cerevisiae, Pichia pastoris, and Schizosaccharomyces pombe. Vectors include integrating, episomal, replicating, centromere, and artificial chromosome plasmids. Vectors are introduced into yeast via transformation or electroporation. Expression is controlled by inducible promoters like GAL or CUP1 in S. cerevisiae and AOX1 in P. pastoris.