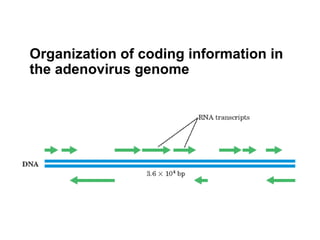
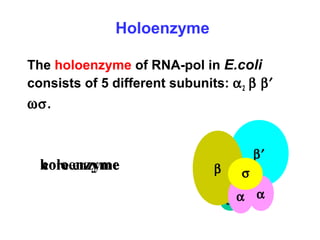
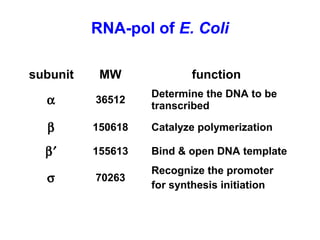
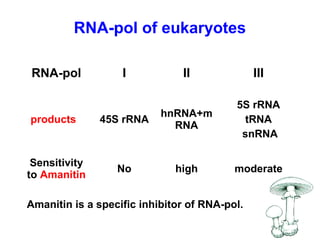
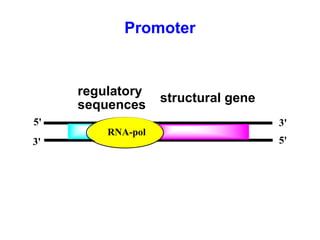
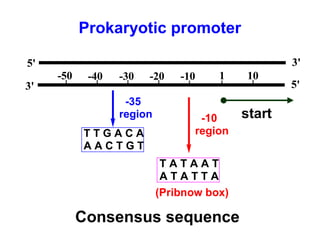
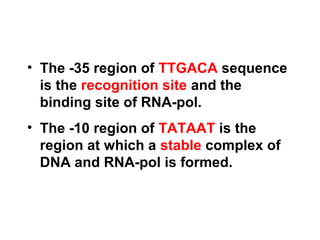
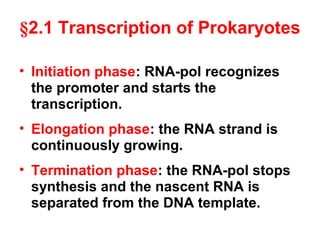
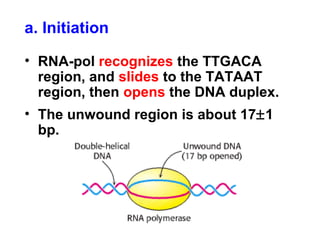
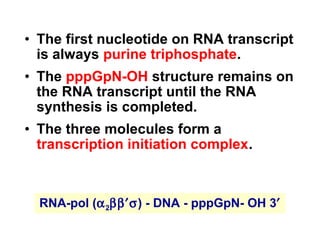
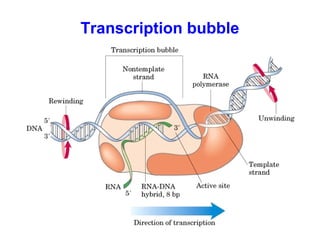
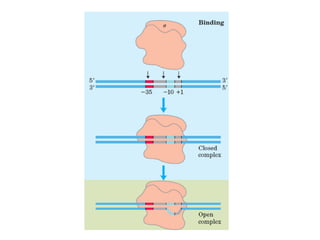
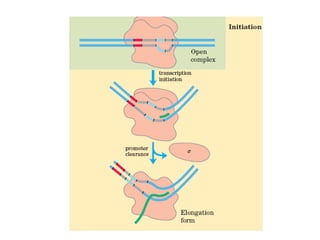
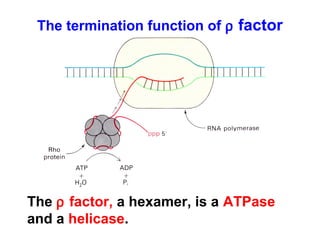
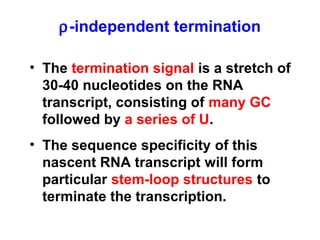
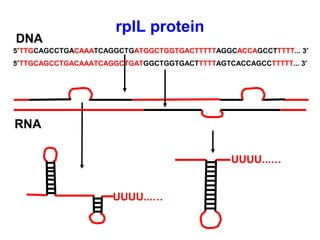
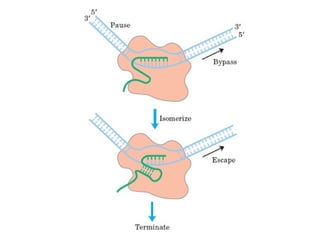
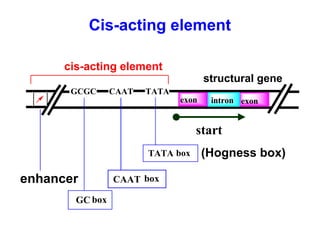
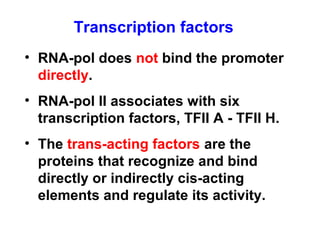
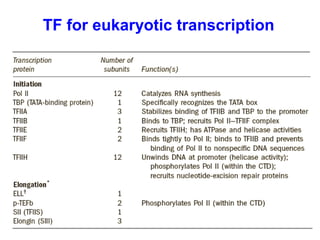
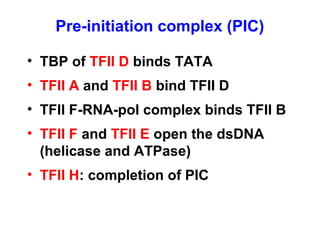
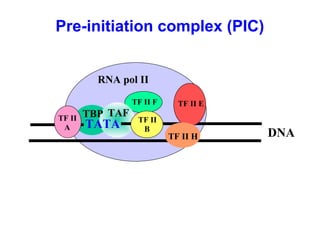
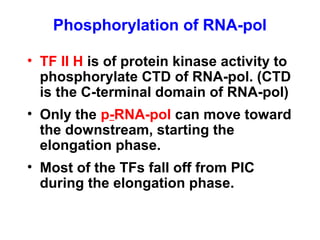
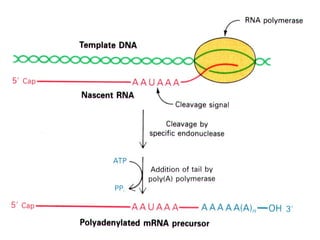
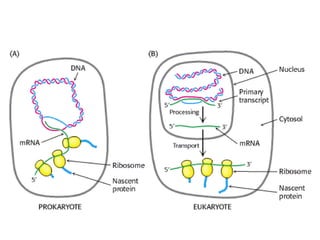
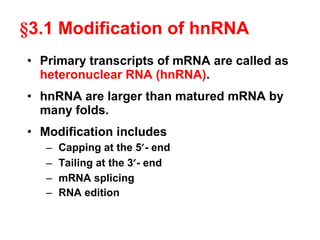
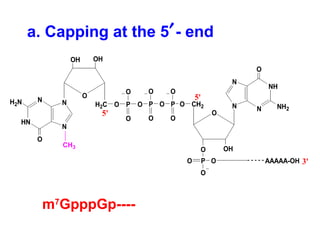
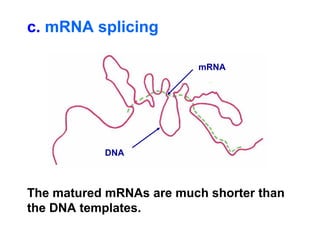
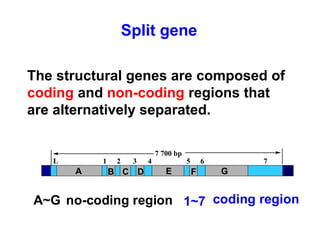
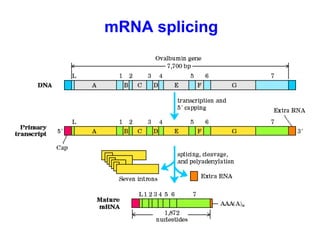
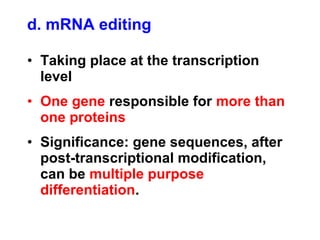
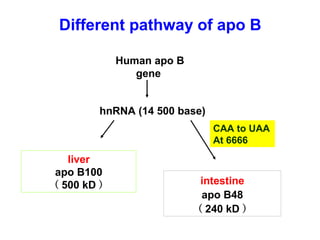
This document summarizes the key processes of transcription in prokaryotes and eukaryotes. It describes how transcription involves initiation, elongation, and termination phases. In prokaryotes, RNA polymerase directly binds DNA and synthesizes RNA from 5' to 3'. In eukaryotes, RNA polymerase requires transcription factors to initiate transcription. The document also discusses post-transcriptional modifications of pre-mRNA in eukaryotes including 5' capping, 3' polyadenylation, splicing of exons and introns, and RNA editing.