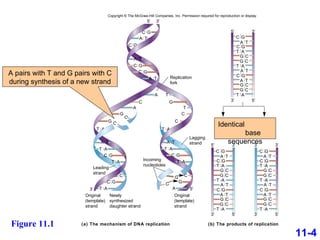
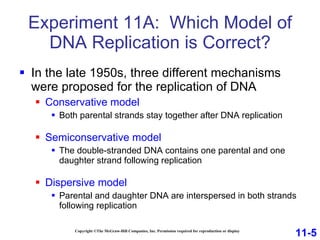
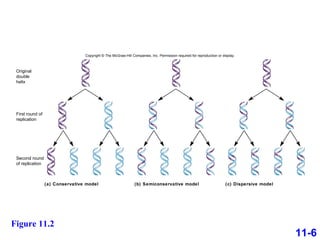
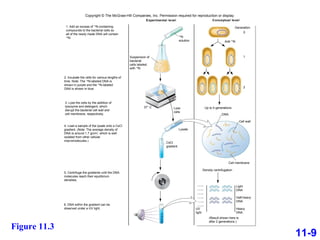
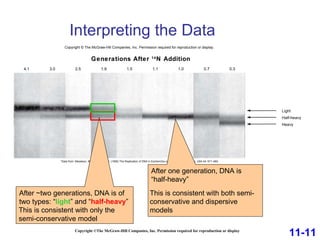
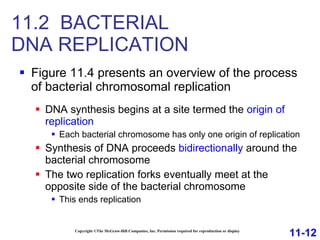
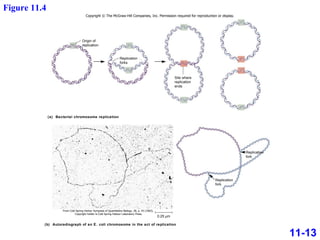
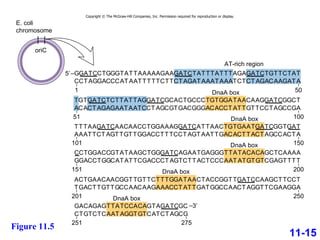
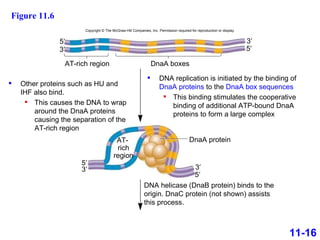
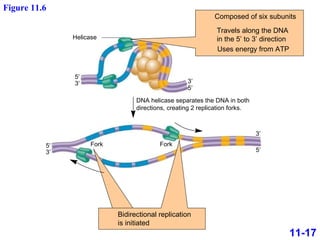
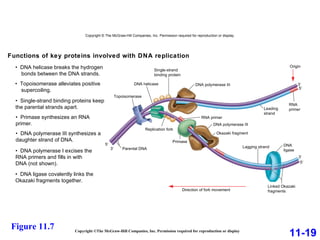
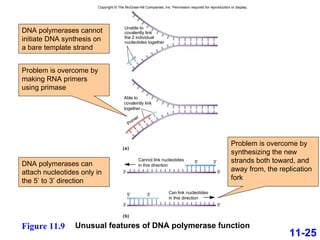
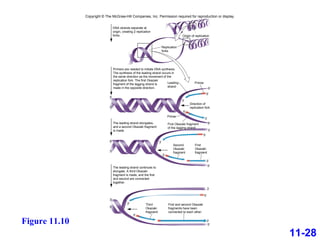
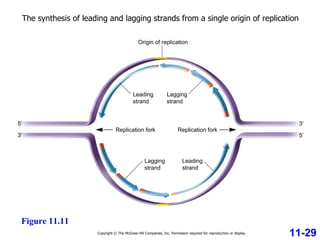
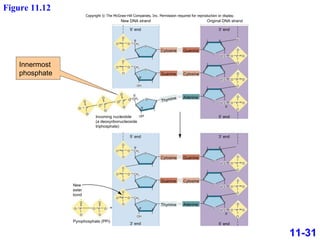
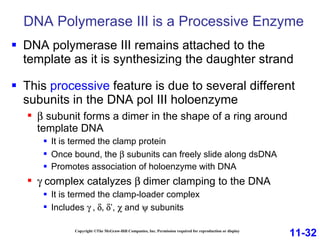
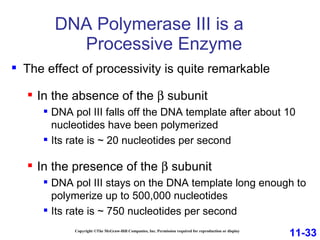
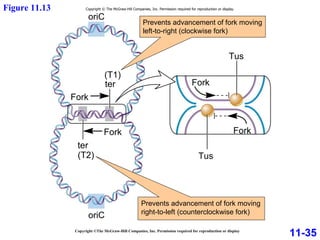
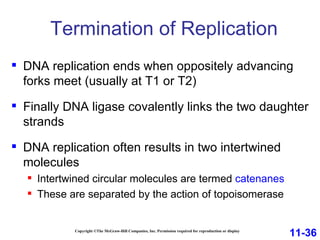
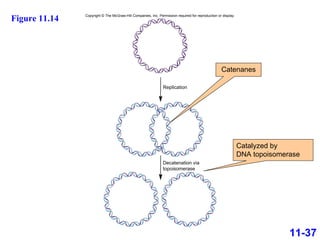
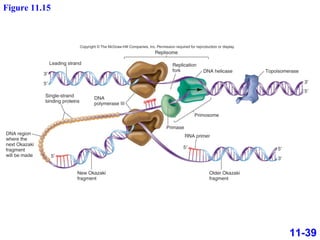
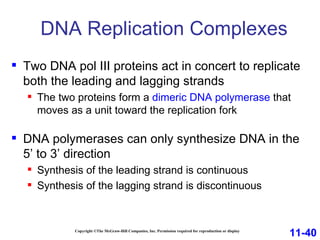
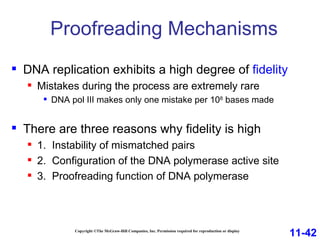
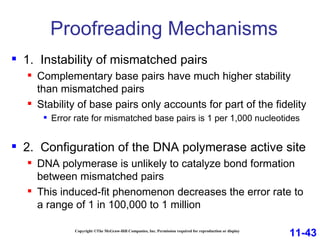
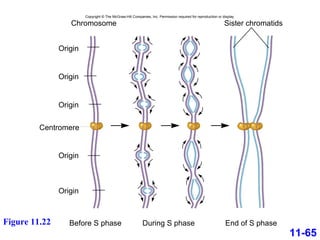
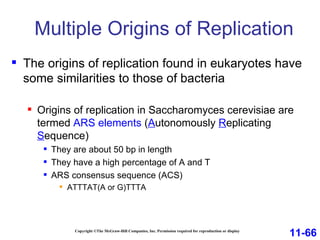
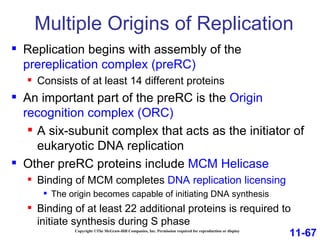
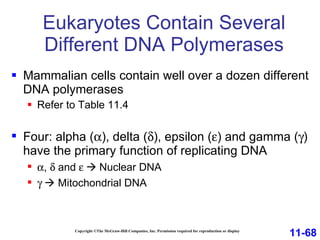
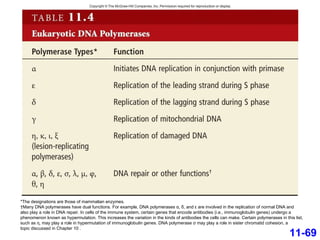
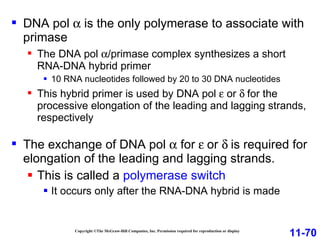
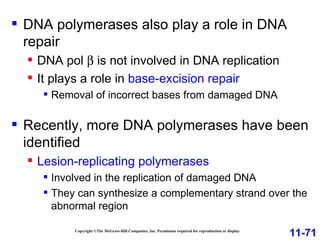
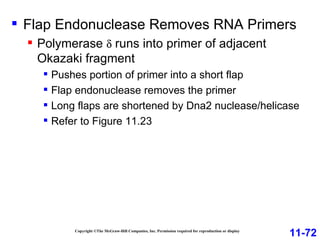
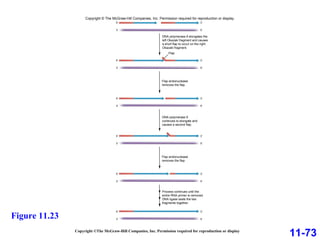
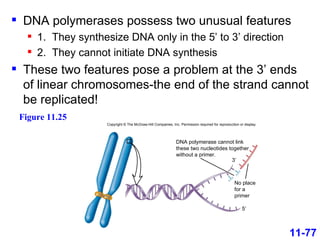
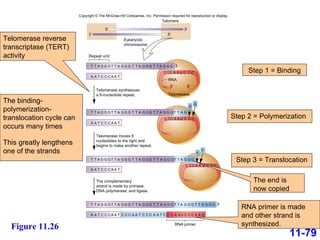
DNA replication is the process by which DNA copies itself. It occurs through semiconservative replication where the two original DNA strands separate and each serves as a template to produce two new daughter strands. On the leading strand, DNA polymerase can continuously synthesize new DNA in the 5' to 3' direction. On the lagging strand, DNA is synthesized away from the replication fork in short fragments called Okazaki fragments that are later joined together.